Steffen Oeltze-Jafra
Auto-nnU-Net: Towards Automated Medical Image Segmentation
May 22, 2025Abstract:Medical Image Segmentation (MIS) includes diverse tasks, from bone to organ segmentation, each with its own challenges in finding the best segmentation model. The state-of-the-art AutoML-related MIS-framework nnU-Net automates many aspects of model configuration but remains constrained by fixed hyperparameters and heuristic design choices. As a full-AutoML framework for MIS, we propose Auto-nnU-Net, a novel nnU-Net variant enabling hyperparameter optimization (HPO), neural architecture search (NAS), and hierarchical NAS (HNAS). Additionally, we propose Regularized PriorBand to balance model accuracy with the computational resources required for training, addressing the resource constraints often faced in real-world medical settings that limit the feasibility of extensive training procedures. We evaluate our approach across diverse MIS datasets from the well-established Medical Segmentation Decathlon, analyzing the impact of AutoML techniques on segmentation performance, computational efficiency, and model design choices. The results demonstrate that our AutoML approach substantially improves the segmentation performance of nnU-Net on 6 out of 10 datasets and is on par on the other datasets while maintaining practical resource requirements. Our code is available at https://github.com/LUH-AI/AutonnUNet.
Artificial Intelligence in Pediatric Echocardiography: Exploring Challenges, Opportunities, and Clinical Applications with Explainable AI and Federated Learning
Nov 15, 2024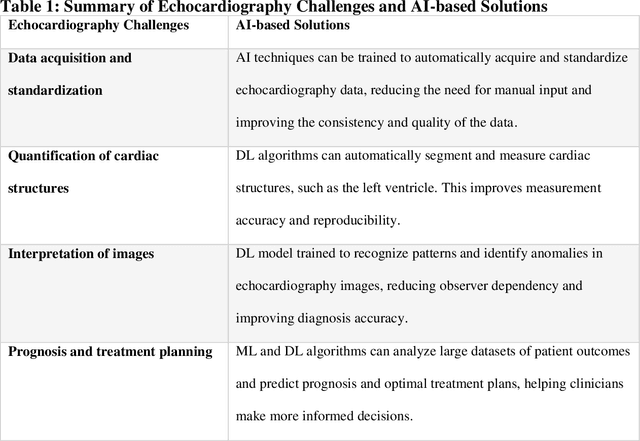
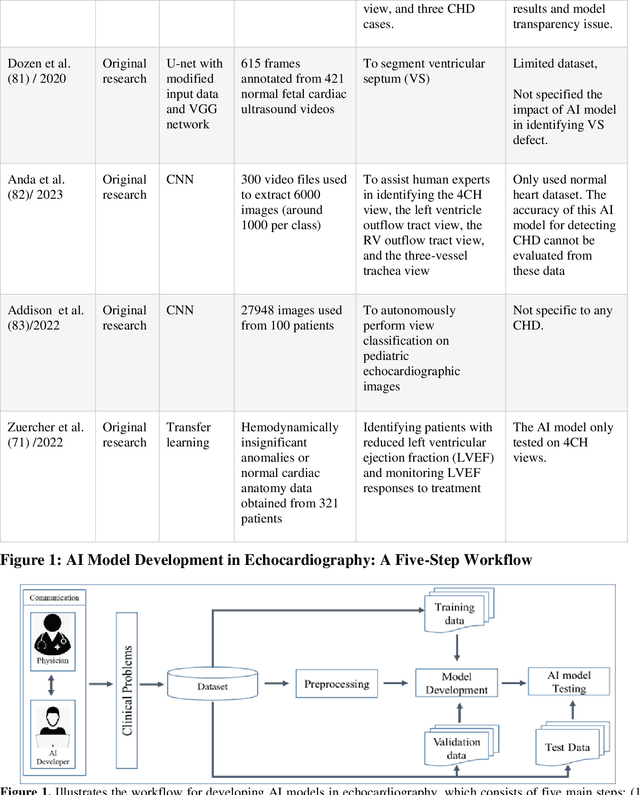


Abstract:Pediatric heart diseases present a broad spectrum of congenital and acquired diseases. More complex congenital malformations require a differentiated and multimodal decision-making process, usually including echocardiography as a central imaging method. Artificial intelligence (AI) offers considerable promise for clinicians by facilitating automated interpretation of pediatric echocardiography data. However, adapting AI technologies for pediatric echocardiography analysis has challenges such as limited public data availability, data privacy, and AI model transparency. Recently, researchers have focused on disruptive technologies, such as federated learning (FL) and explainable AI (XAI), to improve automatic diagnostic and decision support workflows. This study offers a comprehensive overview of the limitations and opportunities of AI in pediatric echocardiography, emphasizing the synergistic workflow and role of XAI and FL, identifying research gaps, and exploring potential future developments. Additionally, three relevant clinical use cases demonstrate the functionality of XAI and FL with a focus on (i) view recognition, (ii) disease classification, (iii) segmentation of cardiac structures, and (iv) quantitative assessment of cardiac function.
MedDoc-Bot: A Chat Tool for Comparative Analysis of Large Language Models in the Context of the Pediatric Hypertension Guideline
May 06, 2024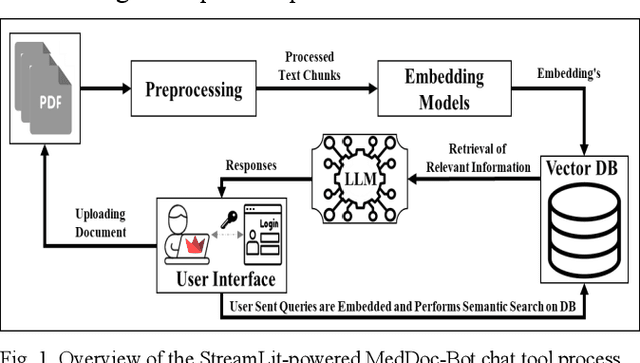
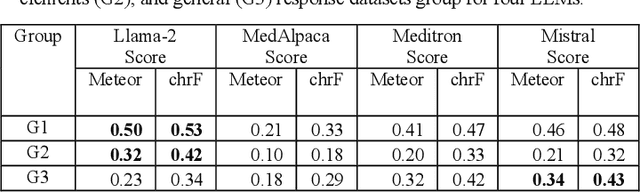
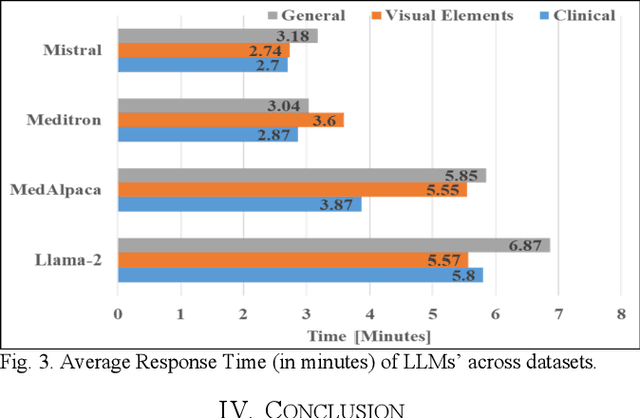
Abstract:This research focuses on evaluating the non-commercial open-source large language models (LLMs) Meditron, MedAlpaca, Mistral, and Llama-2 for their efficacy in interpreting medical guidelines saved in PDF format. As a specific test scenario, we applied these models to the guidelines for hypertension in children and adolescents provided by the European Society of Cardiology (ESC). Leveraging Streamlit, a Python library, we developed a user-friendly medical document chatbot tool (MedDoc-Bot). This tool enables authorized users to upload PDF files and pose questions, generating interpretive responses from four locally stored LLMs. A pediatric expert provides a benchmark for evaluation by formulating questions and responses extracted from the ESC guidelines. The expert rates the model-generated responses based on their fidelity and relevance. Additionally, we evaluated the METEOR and chrF metric scores to assess the similarity of model responses to reference answers. Our study found that Llama-2 and Mistral performed well in metrics evaluation. However, Llama-2 was slower when dealing with text and tabular data. In our human evaluation, we observed that responses created by Mistral, Meditron, and Llama-2 exhibited reasonable fidelity and relevance. This study provides valuable insights into the strengths and limitations of LLMs for future developments in medical document interpretation. Open-Source Code: https://github.com/yaseen28/MedDoc-Bot
Automated SSIM Regression for Detection and Quantification of Motion Artefacts in Brain MR Images
Jun 14, 2022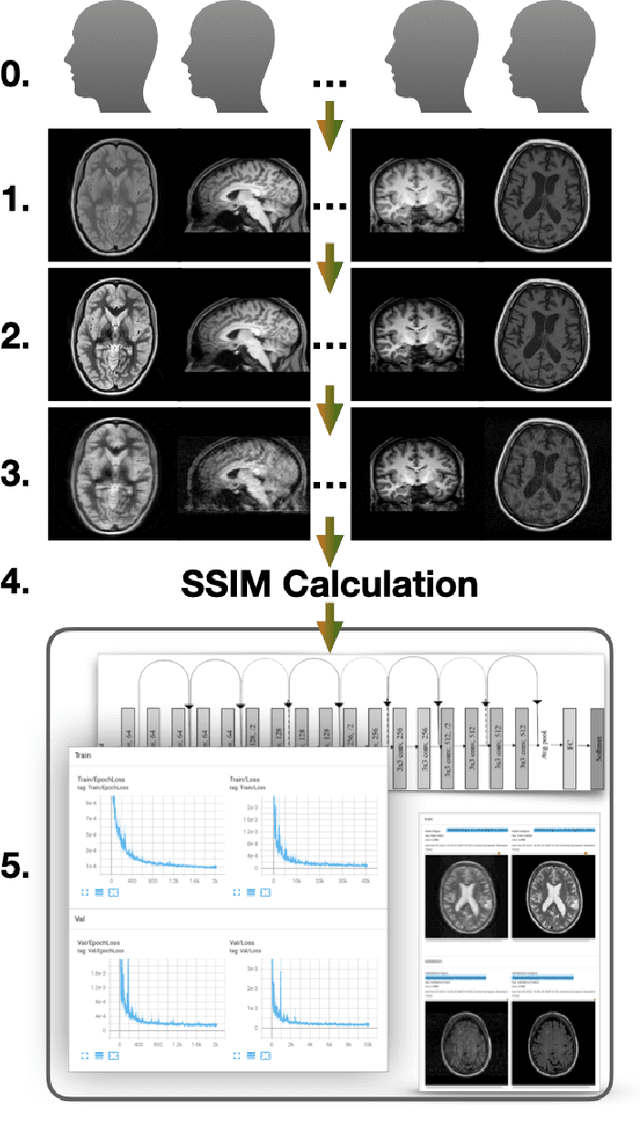
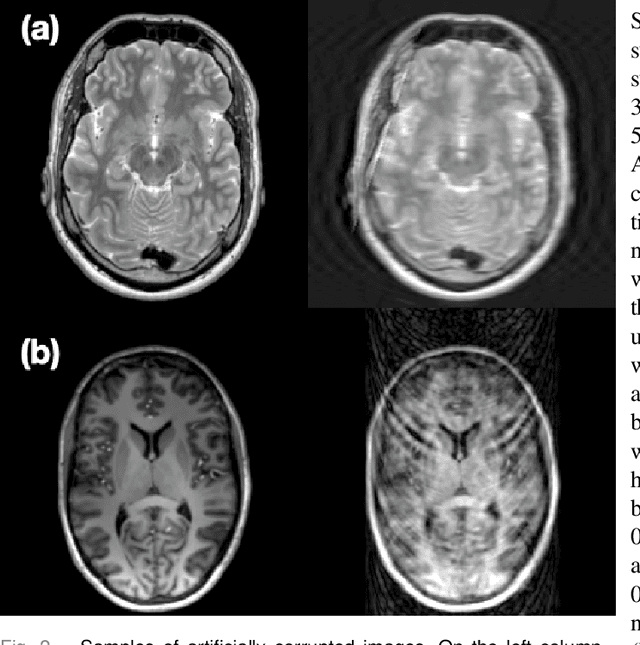
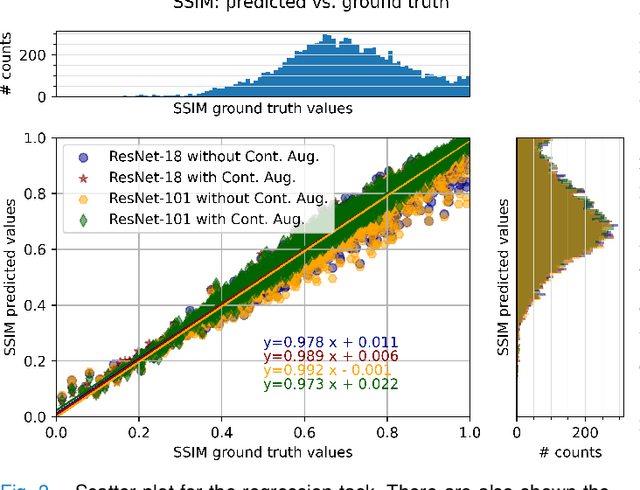
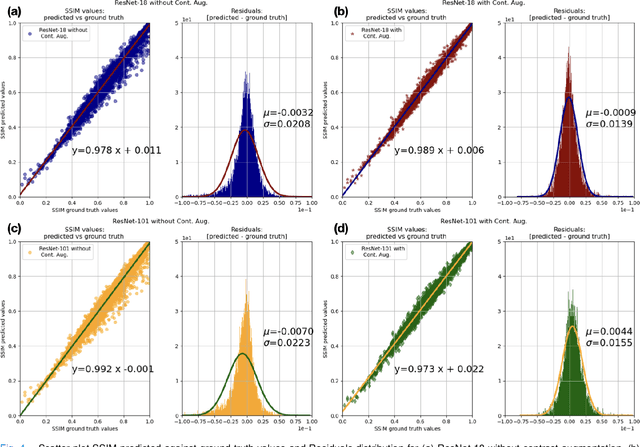
Abstract:Motion artefacts in magnetic resonance brain images are a crucial issue. The assessment of MR image quality is fundamental before proceeding with the clinical diagnosis. If the motion artefacts alter a correct delineation of structure and substructures of the brain, lesions, tumours and so on, the patients need to be re-scanned. Otherwise, neuro-radiologists could report an inaccurate or incorrect diagnosis. The first step right after scanning a patient is the "\textit{image quality assessment}" in order to decide if the acquired images are diagnostically acceptable. An automated image quality assessment based on the structural similarity index (SSIM) regression through a residual neural network has been proposed here, with the possibility to perform also the classification in different groups - by subdividing with SSIM ranges. This method predicts SSIM values of an input image in the absence of a reference ground truth image. The networks were able to detect motion artefacts, and the best performance for the regression and classification task has always been achieved with ResNet-18 with contrast augmentation. Mean and standard deviation of residuals' distribution were $\mu=-0.0009$ and $\sigma=0.0139$, respectively. Whilst for the classification task in 3, 5 and 10 classes, the best accuracies were 97, 95 and 89\%, respectively. The obtained results show that the proposed method could be a tool in supporting neuro-radiologists and radiographers in evaluating the image quality before the diagnosis.
StRegA: Unsupervised Anomaly Detection in Brain MRIs using a Compact Context-encoding Variational Autoencoder
Jan 31, 2022



Abstract:Expert interpretation of anatomical images of the human brain is the central part of neuro-radiology. Several machine learning-based techniques have been proposed to assist in the analysis process. However, the ML models typically need to be trained to perform a specific task, e.g., brain tumour segmentation or classification. Not only do the corresponding training data require laborious manual annotations, but a wide variety of abnormalities can be present in a human brain MRI - even more than one simultaneously, which renders representation of all possible anomalies very challenging. Hence, a possible solution is an unsupervised anomaly detection (UAD) system that can learn a data distribution from an unlabelled dataset of healthy subjects and then be applied to detect out of distribution samples. Such a technique can then be used to detect anomalies - lesions or abnormalities, for example, brain tumours, without explicitly training the model for that specific pathology. Several Variational Autoencoder (VAE) based techniques have been proposed in the past for this task. Even though they perform very well on controlled artificially simulated anomalies, many of them perform poorly while detecting anomalies in clinical data. This research proposes a compact version of the "context-encoding" VAE (ceVAE) model, combined with pre and post-processing steps, creating a UAD pipeline (StRegA), which is more robust on clinical data, and shows its applicability in detecting anomalies such as tumours in brain MRIs. The proposed pipeline achieved a Dice score of 0.642$\pm$0.101 while detecting tumours in T2w images of the BraTS dataset and 0.859$\pm$0.112 while detecting artificially induced anomalies, while the best performing baseline achieved 0.522$\pm$0.135 and 0.783$\pm$0.111, respectively.
ShuffleUNet: Super resolution of diffusion-weighted MRIs using deep learning
Feb 25, 2021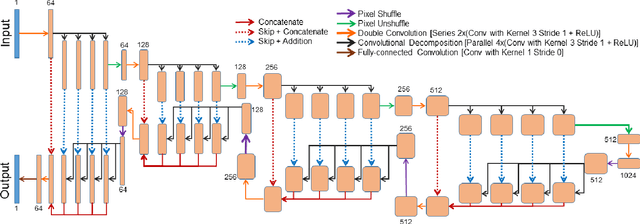
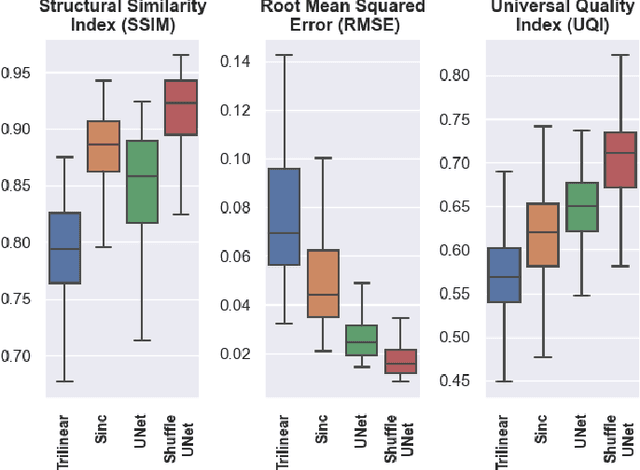
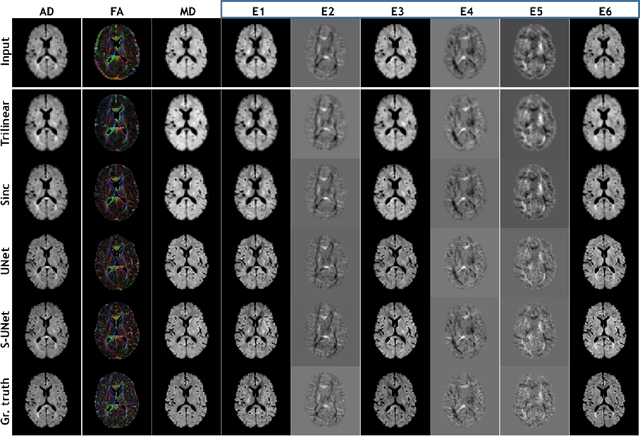
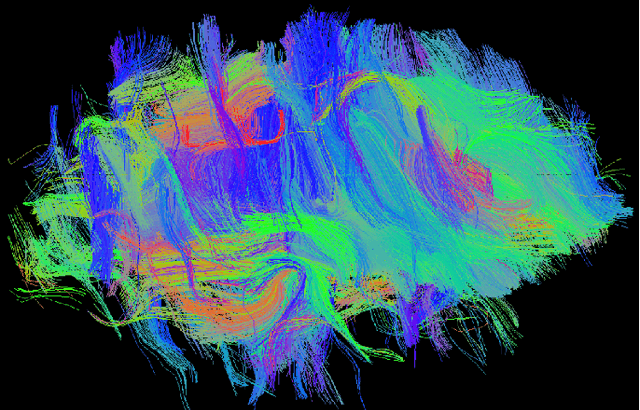
Abstract:Diffusion-weighted magnetic resonance imaging (DW-MRI) can be used to characterise the microstructure of the nervous tissue, e.g. to delineate brain white matter connections in a non-invasive manner via fibre tracking. Magnetic Resonance Imaging (MRI) in high spatial resolution would play an important role in visualising such fibre tracts in a superior manner. However, obtaining an image of such resolution comes at the expense of longer scan time. Longer scan time can be associated with the increase of motion artefacts, due to the patient's psychological and physical conditions. Single Image Super-Resolution (SISR), a technique aimed to obtain high-resolution (HR) details from one single low-resolution (LR) input image, achieved with Deep Learning, is the focus of this study. Compared to interpolation techniques or sparse-coding algorithms, deep learning extracts prior knowledge from big datasets and produces superior MRI images from the low-resolution counterparts. In this research, a deep learning based super-resolution technique is proposed and has been applied for DW-MRI. Images from the IXI dataset have been used as the ground-truth and were artificially downsampled to simulate the low-resolution images. The proposed method has shown statistically significant improvement over the baselines and achieved an SSIM of $0.913\pm0.045$.
Retrospective Motion Correction of MR Images using Prior-Assisted Deep Learning
Nov 28, 2020

Abstract:In MRI, motion artefacts are among the most common types of artefacts. They can degrade images and render them unusable for accurate diagnosis. Traditional methods, such as prospective or retrospective motion correction, have been proposed to avoid or alleviate motion artefacts. Recently, several other methods based on deep learning approaches have been proposed to solve this problem. This work proposes to enhance the performance of existing deep learning models by the inclusion of additional information present as image priors. The proposed approach has shown promising results and will be further investigated for clinical validity.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge