Christoph Reich
Feed-Forward SceneDINO for Unsupervised Semantic Scene Completion
Jul 08, 2025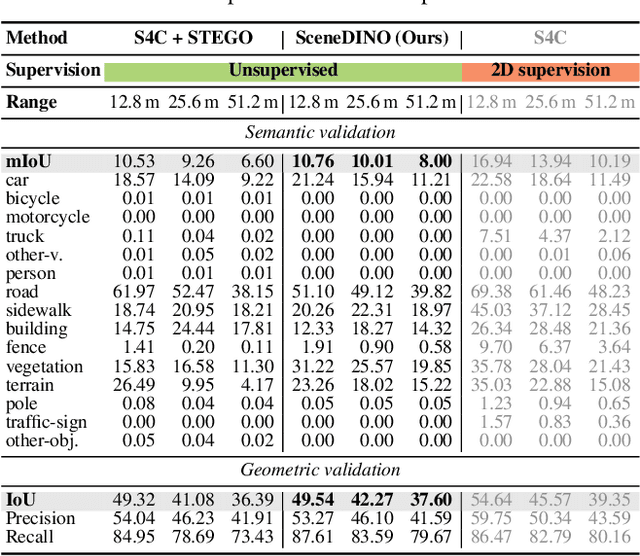
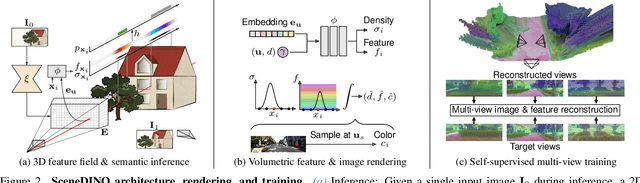

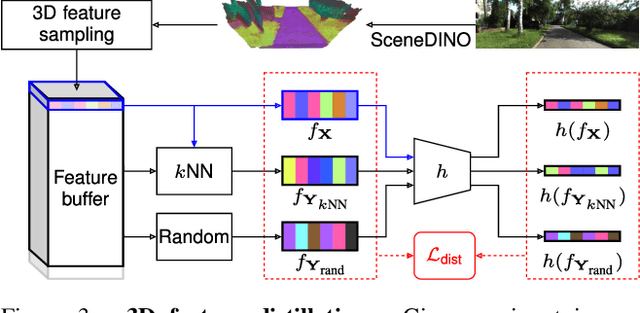
Abstract:Semantic scene completion (SSC) aims to infer both the 3D geometry and semantics of a scene from single images. In contrast to prior work on SSC that heavily relies on expensive ground-truth annotations, we approach SSC in an unsupervised setting. Our novel method, SceneDINO, adapts techniques from self-supervised representation learning and 2D unsupervised scene understanding to SSC. Our training exclusively utilizes multi-view consistency self-supervision without any form of semantic or geometric ground truth. Given a single input image, SceneDINO infers the 3D geometry and expressive 3D DINO features in a feed-forward manner. Through a novel 3D feature distillation approach, we obtain unsupervised 3D semantics. In both 3D and 2D unsupervised scene understanding, SceneDINO reaches state-of-the-art segmentation accuracy. Linear probing our 3D features matches the segmentation accuracy of a current supervised SSC approach. Additionally, we showcase the domain generalization and multi-view consistency of SceneDINO, taking the first steps towards a strong foundation for single image 3D scene understanding.
Scene-Centric Unsupervised Panoptic Segmentation
Apr 02, 2025Abstract:Unsupervised panoptic segmentation aims to partition an image into semantically meaningful regions and distinct object instances without training on manually annotated data. In contrast to prior work on unsupervised panoptic scene understanding, we eliminate the need for object-centric training data, enabling the unsupervised understanding of complex scenes. To that end, we present the first unsupervised panoptic method that directly trains on scene-centric imagery. In particular, we propose an approach to obtain high-resolution panoptic pseudo labels on complex scene-centric data, combining visual representations, depth, and motion cues. Utilizing both pseudo-label training and a panoptic self-training strategy yields a novel approach that accurately predicts panoptic segmentation of complex scenes without requiring any human annotations. Our approach significantly improves panoptic quality, e.g., surpassing the recent state of the art in unsupervised panoptic segmentation on Cityscapes by 9.4% points in PQ.
Arbitrary Data as Images: Fusion of Patient Data Across Modalities and Irregular Intervals with Vision Transformers
Jan 30, 2025



Abstract:A patient undergoes multiple examinations in each hospital stay, where each provides different facets of the health status. These assessments include temporal data with varying sampling rates, discrete single-point measurements, therapeutic interventions such as medication administration, and images. While physicians are able to process and integrate diverse modalities intuitively, neural networks need specific modeling for each modality complicating the training procedure. We demonstrate that this complexity can be significantly reduced by visualizing all information as images along with unstructured text and subsequently training a conventional vision-text transformer. Our approach, Vision Transformer for irregular sampled Multi-modal Measurements (ViTiMM), not only simplifies data preprocessing and modeling but also outperforms current state-of-the-art methods in predicting in-hospital mortality and phenotyping, as evaluated on 6,175 patients from the MIMIC-IV dataset. The modalities include patient's clinical measurements, medications, X-ray images, and electrocardiography scans. We hope our work inspires advancements in multi-modal medical AI by reducing the training complexity to (visual) prompt engineering, thus lowering entry barriers and enabling no-code solutions for training. The source code will be made publicly available.
A Perspective on Deep Vision Performance with Standard Image and Video Codecs
Apr 18, 2024
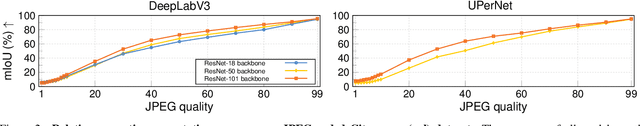
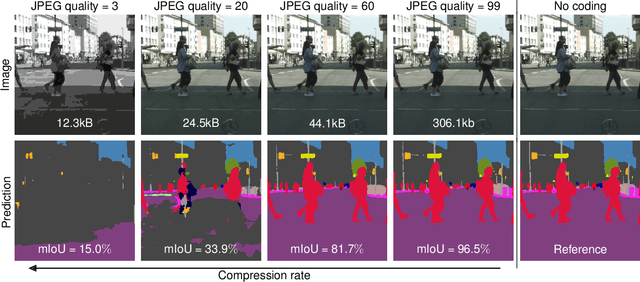
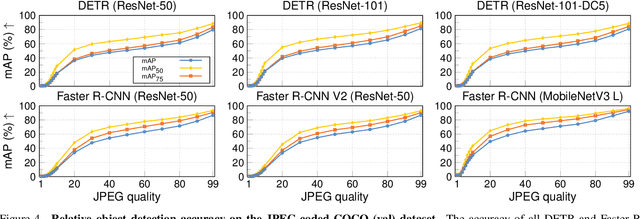
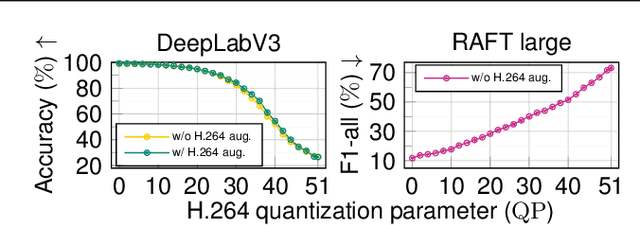
Abstract:Resource-constrained hardware, such as edge devices or cell phones, often rely on cloud servers to provide the required computational resources for inference in deep vision models. However, transferring image and video data from an edge or mobile device to a cloud server requires coding to deal with network constraints. The use of standardized codecs, such as JPEG or H.264, is prevalent and required to ensure interoperability. This paper aims to examine the implications of employing standardized codecs within deep vision pipelines. We find that using JPEG and H.264 coding significantly deteriorates the accuracy across a broad range of vision tasks and models. For instance, strong compression rates reduce semantic segmentation accuracy by more than 80% in mIoU. In contrast to previous findings, our analysis extends beyond image and action classification to localization and dense prediction tasks, thus providing a more comprehensive perspective.
Deep Video Codec Control
Sep 16, 2023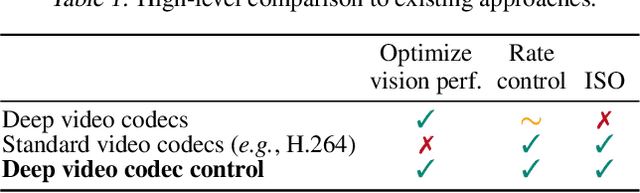
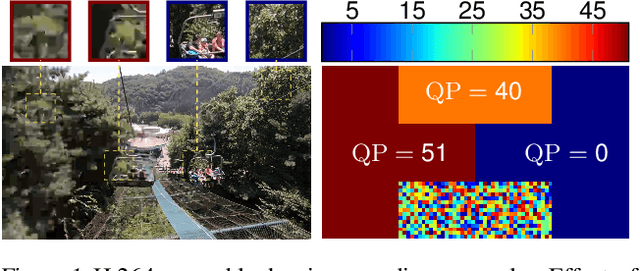
Abstract:Lossy video compression is commonly used when transmitting and storing video data. Unified video codecs (e.g., H.264 or H.265) remain the de facto standard, despite the availability of advanced (neural) compression approaches. Transmitting videos in the face of dynamic network bandwidth conditions requires video codecs to adapt to vastly different compression strengths. Rate control modules augment the codec's compression such that bandwidth constraints are satisfied and video distortion is minimized. While, both standard video codes and their rate control modules are developed to minimize video distortion w.r.t. human quality assessment, preserving the downstream performance of deep vision models is not considered. In this paper, we present the first end-to-end learnable deep video codec control considering both bandwidth constraints and downstream vision performance, while not breaking existing standardization. We demonstrate for two common vision tasks (semantic segmentation and optical flow estimation) and on two different datasets that our deep codec control better preserves downstream performance than using 2-pass average bit rate control while meeting dynamic bandwidth constraints and adhering to standardizations.
Differentiable JPEG: The Devil is in the Details
Sep 13, 2023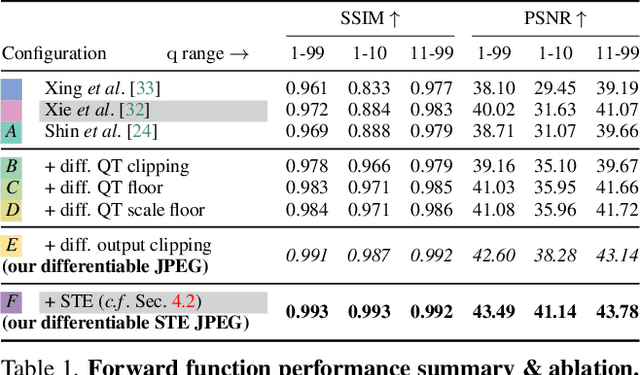
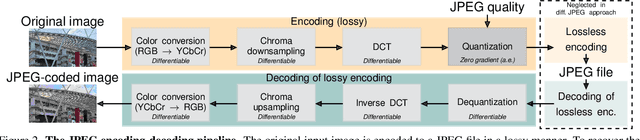
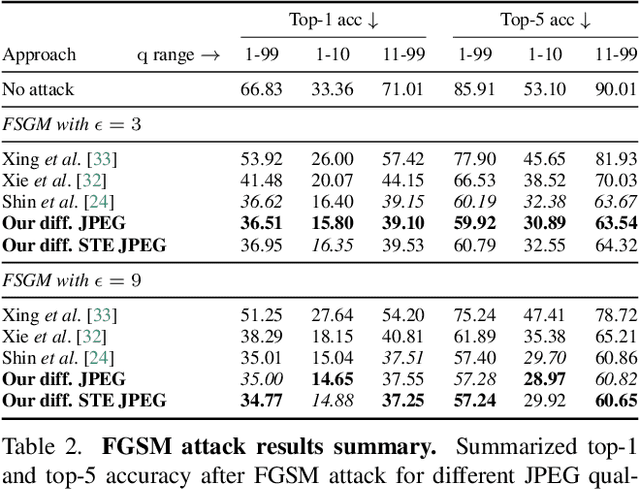
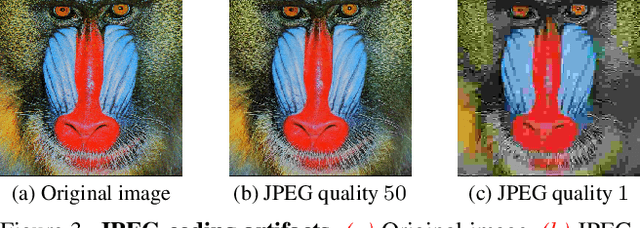
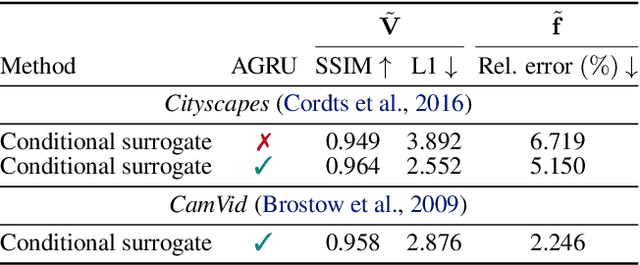
Abstract:JPEG remains one of the most widespread lossy image coding methods. However, the non-differentiable nature of JPEG restricts the application in deep learning pipelines. Several differentiable approximations of JPEG have recently been proposed to address this issue. This paper conducts a comprehensive review of existing diff. JPEG approaches and identifies critical details that have been missed by previous methods. To this end, we propose a novel diff. JPEG approach, overcoming previous limitations. Our approach is differentiable w.r.t. the input image, the JPEG quality, the quantization tables, and the color conversion parameters. We evaluate the forward and backward performance of our diff. JPEG approach against existing methods. Additionally, extensive ablations are performed to evaluate crucial design choices. Our proposed diff. JPEG resembles the (non-diff.) reference implementation best, significantly surpassing the recent-best diff. approach by $3.47$dB (PSNR) on average. For strong compression rates, we can even improve PSNR by $9.51$dB. Strong adversarial attack results are yielded by our diff. JPEG, demonstrating the effective gradient approximation. Our code is available at https://github.com/necla-ml/Diff-JPEG.
The TYC Dataset for Understanding Instance-Level Semantics and Motions of Cells in Microstructures
Aug 23, 2023
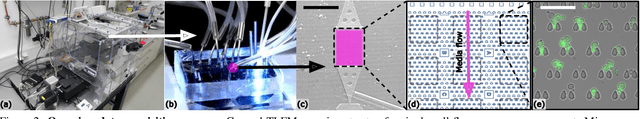

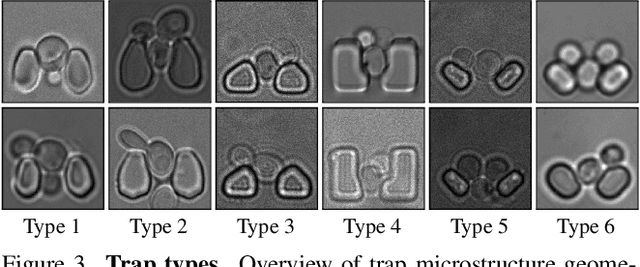
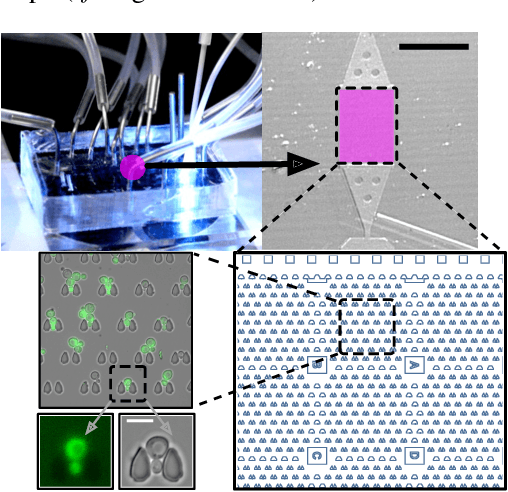
Abstract:Segmenting cells and tracking their motion over time is a common task in biomedical applications. However, predicting accurate instance-wise segmentation and cell motions from microscopy imagery remains a challenging task. Using microstructured environments for analyzing single cells in a constant flow of media adds additional complexity. While large-scale labeled microscopy datasets are available, we are not aware of any large-scale dataset, including both cells and microstructures. In this paper, we introduce the trapped yeast cell (TYC) dataset, a novel dataset for understanding instance-level semantics and motions of cells in microstructures. We release $105$ dense annotated high-resolution brightfield microscopy images, including about $19$k instance masks. We also release $261$ curated video clips composed of $1293$ high-resolution microscopy images to facilitate unsupervised understanding of cell motions and morphology. TYC offers ten times more instance annotations than the previously largest dataset, including cells and microstructures. Our effort also exceeds previous attempts in terms of microstructure variability, resolution, complexity, and capturing device (microscopy) variability. We facilitate a unified comparison on our novel dataset by introducing a standardized evaluation strategy. TYC and evaluation code are publicly available under CC BY 4.0 license.
An Instance Segmentation Dataset of Yeast Cells in Microstructures
Apr 23, 2023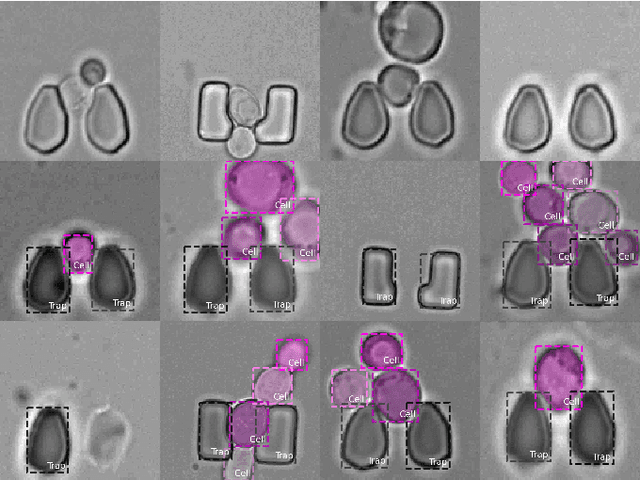
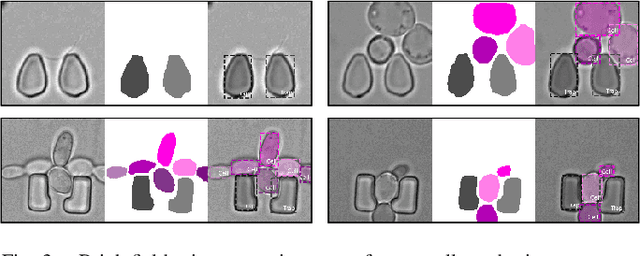
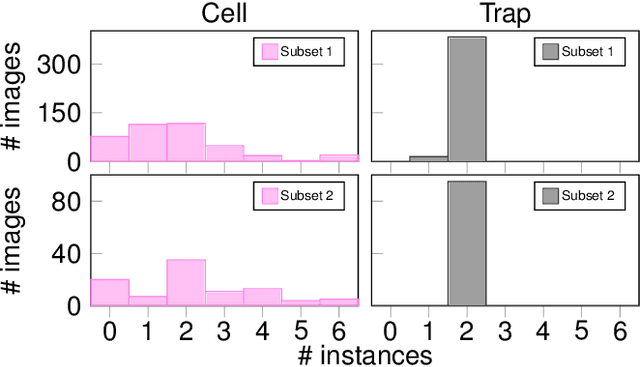
Abstract:Extracting single-cell information from microscopy data requires accurate instance-wise segmentations. Obtaining pixel-wise segmentations from microscopy imagery remains a challenging task, especially with the added complexity of microstructured environments. This paper presents a novel dataset for segmenting yeast cells in microstructures. We offer pixel-wise instance segmentation labels for both cells and trap microstructures. In total, we release 493 densely annotated microscopy images. To facilitate a unified comparison between novel segmentation algorithms, we propose a standardized evaluation strategy for our dataset. The aim of the dataset and evaluation strategy is to facilitate the development of new cell segmentation approaches. The dataset is publicly available at https://christophreich1996.github.io/yeast_in_microstructures_dataset/ .
Histopathological Image Classification based on Self-Supervised Vision Transformer and Weak Labels
Oct 17, 2022Abstract:Whole Slide Image (WSI) analysis is a powerful method to facilitate the diagnosis of cancer in tissue samples. Automating this diagnosis poses various issues, most notably caused by the immense image resolution and limited annotations. WSIs commonly exhibit resolutions of 100Kx100K pixels. Annotating cancerous areas in WSIs on the pixel level is prohibitively labor-intensive and requires a high level of expert knowledge. Multiple instance learning (MIL) alleviates the need for expensive pixel-level annotations. In MIL, learning is performed on slide-level labels, in which a pathologist provides information about whether a slide includes cancerous tissue. Here, we propose Self-ViT-MIL, a novel approach for classifying and localizing cancerous areas based on slide-level annotations, eliminating the need for pixel-wise annotated training data. Self-ViT- MIL is pre-trained in a self-supervised setting to learn rich feature representation without relying on any labels. The recent Vision Transformer (ViT) architecture builds the feature extractor of Self-ViT-MIL. For localizing cancerous regions, a MIL aggregator with global attention is utilized. To the best of our knowledge, Self-ViT- MIL is the first approach to introduce self-supervised ViTs in MIL-based WSI analysis tasks. We showcase the effectiveness of our approach on the common Camelyon16 dataset. Self-ViT-MIL surpasses existing state-of-the-art MIL-based approaches in terms of accuracy and area under the curve (AUC).
Scalable 3D Semantic Segmentation for Gun Detection in CT Scans
Dec 07, 2021
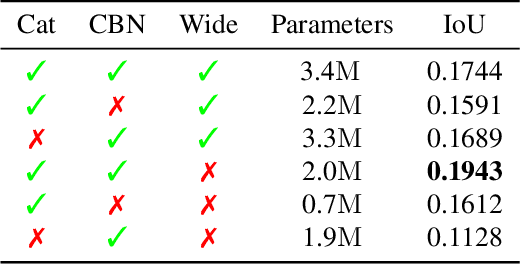
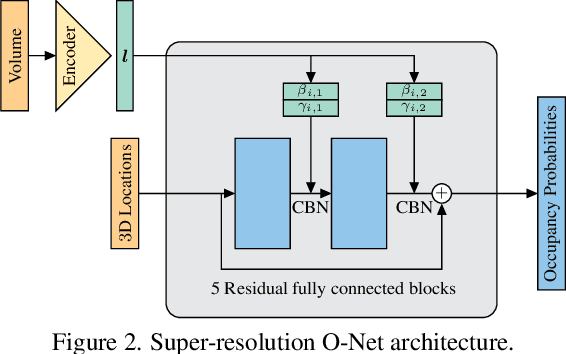
Abstract:With the increased availability of 3D data, the need for solutions processing those also increased rapidly. However, adding dimension to already reliably accurate 2D approaches leads to immense memory consumption and higher computational complexity. These issues cause current hardware to reach its limitations, with most methods forced to reduce the input resolution drastically. Our main contribution is a novel deep 3D semantic segmentation method for gun detection in baggage CT scans that enables fast training and low video memory consumption for high-resolution voxelized volumes. We introduce a moving pyramid approach that utilizes multiple forward passes at inference time for segmenting an instance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge