André O. Françani
Motion Consistency Loss for Monocular Visual Odometry with Attention-Based Deep Learning
Jan 19, 2024Abstract:Deep learning algorithms have driven expressive progress in many complex tasks. The loss function is a core component of deep learning techniques, guiding the learning process of neural networks. This paper contributes by introducing a consistency loss for visual odometry with deep learning-based approaches. The motion consistency loss explores repeated motions that appear in consecutive overlapped video clips. Experimental results show that our approach increased the performance of a model on the KITTI odometry benchmark.
Transformer-based model for monocular visual odometry: a video understanding approach
May 10, 2023

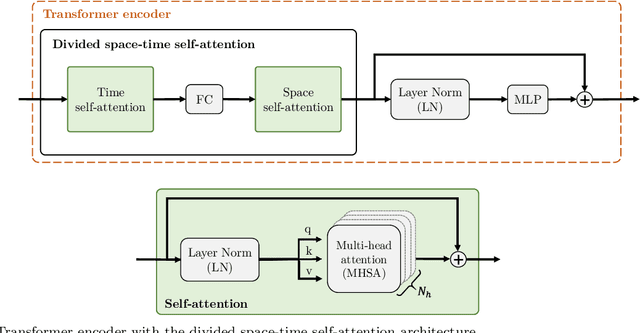

Abstract:Estimating the camera pose given images of a single camera is a traditional task in mobile robots and autonomous vehicles. This problem is called monocular visual odometry and it often relies on geometric approaches that require engineering effort for a specific scenario. Deep learning methods have shown to be generalizable after proper training and a considerable amount of available data. Transformer-based architectures have dominated the state-of-the-art in natural language processing and computer vision tasks, such as image and video understanding. In this work, we deal with the monocular visual odometry as a video understanding task to estimate the 6-DoF camera's pose. We contribute by presenting the TSformer-VO model based on spatio-temporal self-attention mechanisms to extract features from clips and estimate the motions in an end-to-end manner. Our approach achieved competitive state-of-the-art performance compared with geometry-based and deep learning-based methods on the KITTI visual odometry dataset, outperforming the DeepVO implementation highly accepted in the visual odometry community.
An Instance Segmentation Dataset of Yeast Cells in Microstructures
Apr 23, 2023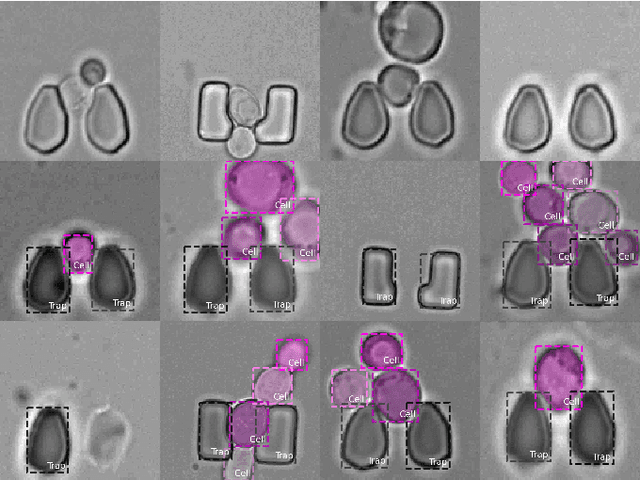
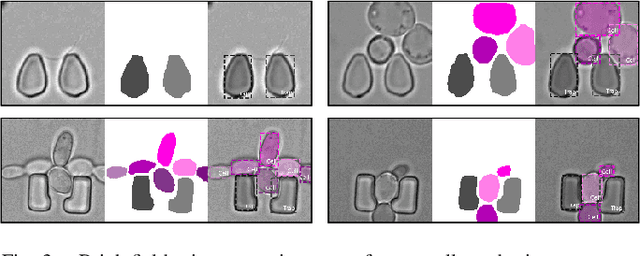
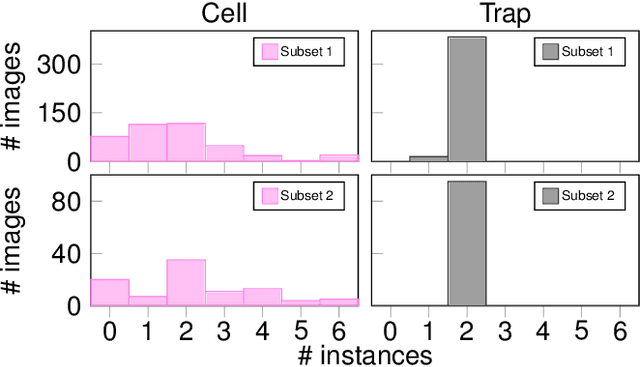
Abstract:Extracting single-cell information from microscopy data requires accurate instance-wise segmentations. Obtaining pixel-wise segmentations from microscopy imagery remains a challenging task, especially with the added complexity of microstructured environments. This paper presents a novel dataset for segmenting yeast cells in microstructures. We offer pixel-wise instance segmentation labels for both cells and trap microstructures. In total, we release 493 densely annotated microscopy images. To facilitate a unified comparison between novel segmentation algorithms, we propose a standardized evaluation strategy for our dataset. The aim of the dataset and evaluation strategy is to facilitate the development of new cell segmentation approaches. The dataset is publicly available at https://christophreich1996.github.io/yeast_in_microstructures_dataset/ .
Analysis of the performance of U-Net neural networks for the segmentation of living cells
Oct 04, 2022



Abstract:The automated analysis of microscopy images is a challenge in the context of single-cell tracking and quantification. This work has as goals the study of the performance of deep learning for segmenting microscopy images and the improvement of the previously available pipeline for tracking single cells. Deep learning techniques, mainly convolutional neural networks, have been applied to cell segmentation problems and have shown high accuracy and fast performance. To perform the image segmentation, an analysis of hyperparameters was done in order to implement a convolutional neural network with U-Net architecture. Furthermore, different models were built in order to optimize the size of the network and the number of learnable parameters. The trained network is then used in the pipeline that localizes the traps in a microfluidic device, performs the image segmentation on trap images, and evaluates the fluorescence intensity and the area of single cells over time. The tracking of the cells during an experiment is performed by image processing algorithms, such as centroid estimation and watershed. Finally, with all improvements in the neural network to segment single cells and in the pipeline, quasi-real-time image analysis was enabled, where 6.20GB of data was processed in 4 minutes.
Dense Prediction Transformer for Scale Estimation in Monocular Visual Odometry
Oct 04, 2022

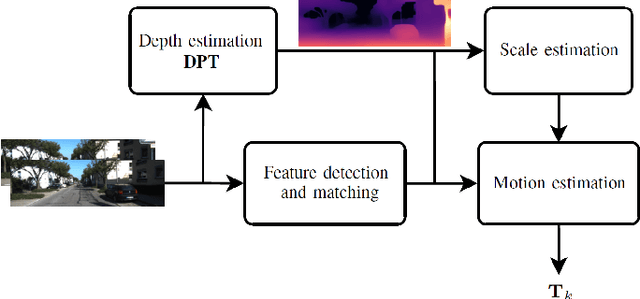
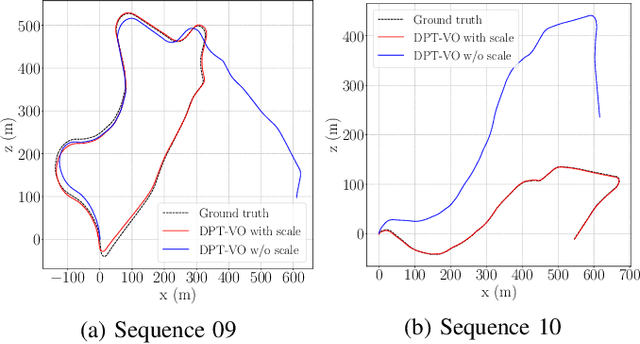
Abstract:Monocular visual odometry consists of the estimation of the position of an agent through images of a single camera, and it is applied in autonomous vehicles, medical robots, and augmented reality. However, monocular systems suffer from the scale ambiguity problem due to the lack of depth information in 2D frames. This paper contributes by showing an application of the dense prediction transformer model for scale estimation in monocular visual odometry systems. Experimental results show that the scale drift problem of monocular systems can be reduced through the accurate estimation of the depth map by this model, achieving competitive state-of-the-art performance on a visual odometry benchmark.
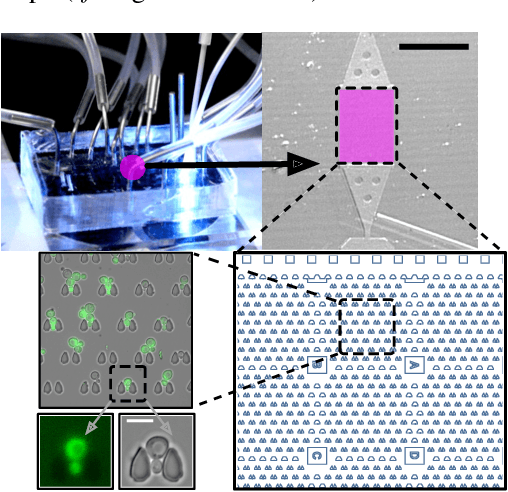
Multiclass Yeast Segmentation in Microstructured Environments with Deep Learning
Nov 19, 2020
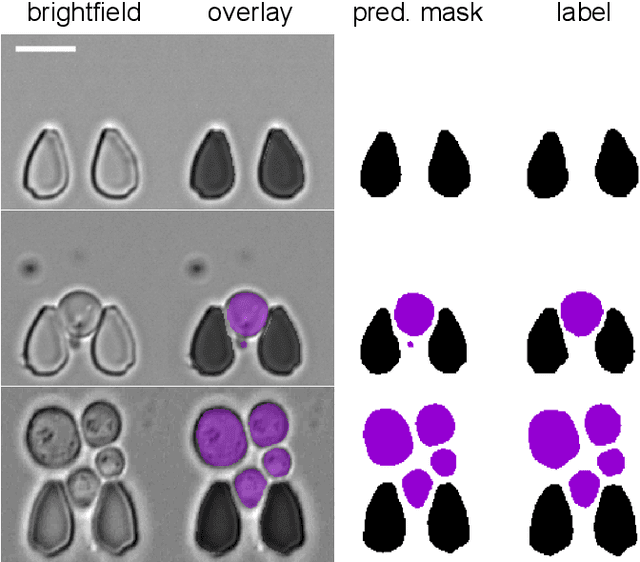
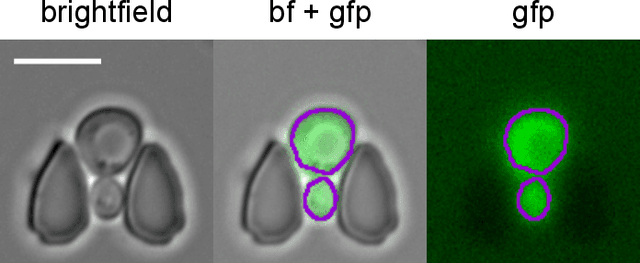

Abstract:Cell segmentation is a major bottleneck in extracting quantitative single-cell information from microscopy data. The challenge is exasperated in the setting of microstructured environments. While deep learning approaches have proven useful for general cell segmentation tasks, existing segmentation tools for the yeast-microstructure setting rely on traditional machine learning approaches. Here we present convolutional neural networks trained for multiclass segmenting of individual yeast cells and discerning these from cell-similar microstructures. We give an overview of the datasets recorded for training, validating and testing the networks, as well as a typical use-case. We showcase the method's contribution to segmenting yeast in microstructured environments with a typical synthetic biology application in mind. The models achieve robust segmentation results, outperforming the previous state-of-the-art in both accuracy and speed. The combination of fast and accurate segmentation is not only beneficial for a posteriori data processing, it also makes online monitoring of thousands of trapped cells or closed-loop optimal experimental design feasible from an image processing perspective.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge