Christian Wildner
Multi-StyleGAN: Towards Image-Based Simulation of Time-Lapse Live-Cell Microscopy
Jun 15, 2021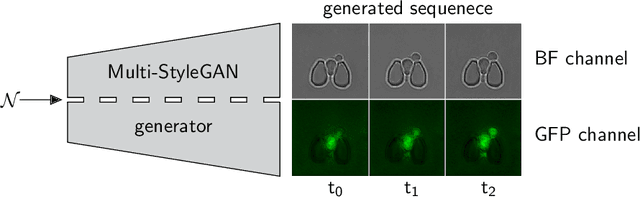

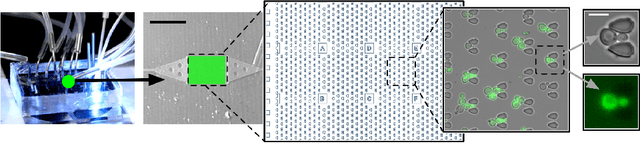
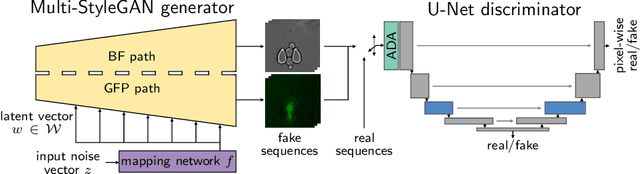
Abstract:Time-lapse fluorescent microscopy (TLFM) combined with predictive mathematical modelling is a powerful tool to study the inherently dynamic processes of life on the single-cell level. Such experiments are costly, complex and labour intensive. A complimentary approach and a step towards completely in silico experiments, is to synthesise the imagery itself. Here, we propose Multi-StyleGAN as a descriptive approach to simulate time-lapse fluorescence microscopy imagery of living cells, based on a past experiment. This novel generative adversarial network synthesises a multi-domain sequence of consecutive timesteps. We showcase Multi-StyleGAN on imagery of multiple live yeast cells in microstructured environments and train on a dataset recorded in our laboratory. The simulation captures underlying biophysical factors and time dependencies, such as cell morphology, growth, physical interactions, as well as the intensity of a fluorescent reporter protein. An immediate application is to generate additional training and validation data for feature extraction algorithms or to aid and expedite development of advanced experimental techniques such as online monitoring or control of cells. Code and dataset is available at https://git.rwth-aachen.de/bcs/projects/tp/multi-stylegan.
Moment-Based Variational Inference for Stochastic Differential Equations
Mar 01, 2021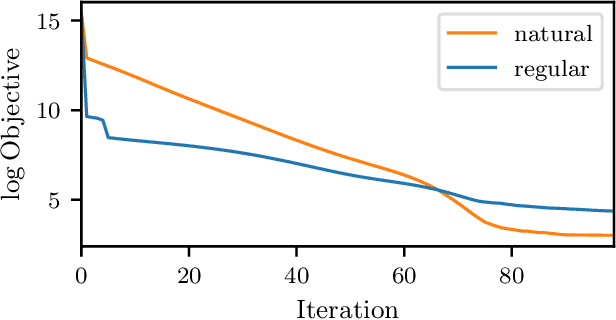



Abstract:Existing deterministic variational inference approaches for diffusion processes use simple proposals and target the marginal density of the posterior. We construct the variational process as a controlled version of the prior process and approximate the posterior by a set of moment functions. In combination with moment closure, the smoothing problem is reduced to a deterministic optimal control problem. Exploiting the path-wise Fisher information, we propose an optimization procedure that corresponds to a natural gradient descent in the variational parameters. Our approach allows for richer variational approximations that extend to state-dependent diffusion terms. The classical Gaussian process approximation is recovered as a special case.
Multiclass Yeast Segmentation in Microstructured Environments with Deep Learning
Nov 19, 2020
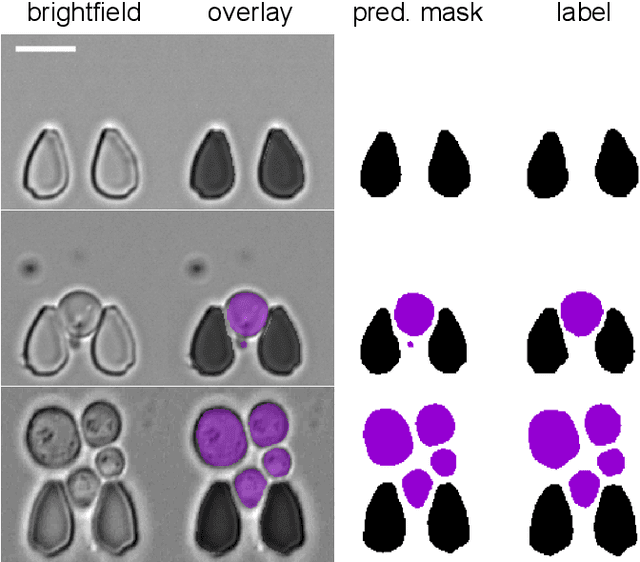
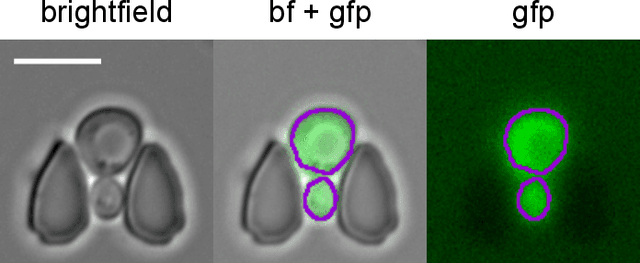

Abstract:Cell segmentation is a major bottleneck in extracting quantitative single-cell information from microscopy data. The challenge is exasperated in the setting of microstructured environments. While deep learning approaches have proven useful for general cell segmentation tasks, existing segmentation tools for the yeast-microstructure setting rely on traditional machine learning approaches. Here we present convolutional neural networks trained for multiclass segmenting of individual yeast cells and discerning these from cell-similar microstructures. We give an overview of the datasets recorded for training, validating and testing the networks, as well as a typical use-case. We showcase the method's contribution to segmenting yeast in microstructured environments with a typical synthetic biology application in mind. The models achieve robust segmentation results, outperforming the previous state-of-the-art in both accuracy and speed. The combination of fast and accurate segmentation is not only beneficial for a posteriori data processing, it also makes online monitoring of thousands of trapped cells or closed-loop optimal experimental design feasible from an image processing perspective.
Moment-Based Variational Inference for Markov Jump Processes
May 14, 2019


Abstract:We propose moment-based variational inference as a flexible framework for approximate smoothing of latent Markov jump processes. The main ingredient of our approach is to partition the set of all transitions of the latent process into classes. This allows to express the Kullback-Leibler divergence between the approximate and the exact posterior process in terms of a set of moment functions that arise naturally from the chosen partition. To illustrate possible choices of the partition, we consider special classes of jump processes that frequently occur in applications. We then extend the results to parameter inference and demonstrate the method on several examples.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge