Tobias Ortmaier
Safe Collision and Clamping Reaction for Parallel Robots During Human-Robot Collaboration
Aug 18, 2023Abstract:Parallel robots (PRs) offer the potential for safe human-robot collaboration because of their low moving masses. Due to the in-parallel kinematic chains, the risk of contact in the form of collisions and clamping at a chain increases. Ensuring safety is investigated in this work through various contact reactions on a real planar PR. External forces are estimated based on proprioceptive information and a dynamics model, which allows contact detection. Retraction along the direction of the estimated line of action provides an instantaneous response to limit the occurring contact forces within the experiment to 70N at a maximum velocity 0.4m/s. A reduction in the stiffness of a Cartesian impedance control is investigated as a further strategy. For clamping, a feedforward neural network (FNN) is trained and tested in different joint angle configurations to classify whether a collision or clamping occurs with an accuracy of 80%. A second FNN classifies the clamping kinematic chain to enable a subsequent kinematic projection of the clamping joint angle onto the rotational platform coordinates. In this way, a structure opening is performed in addition to the softer retraction movement. The reaction strategies are compared in real-world experiments at different velocities and controller stiffnesses to demonstrate their effectiveness. The results show that in all collision and clamping experiments the PR terminates the contact in less than 130ms.
Towards Human-Robot Collaboration with Parallel Robots by Kinetostatic Analysis, Impedance Control and Contact Detection
Aug 18, 2023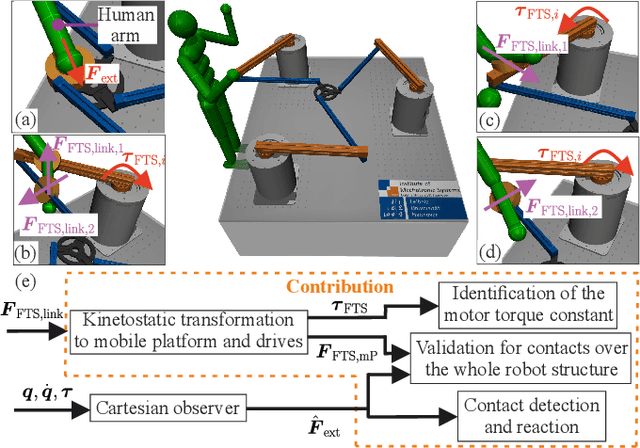
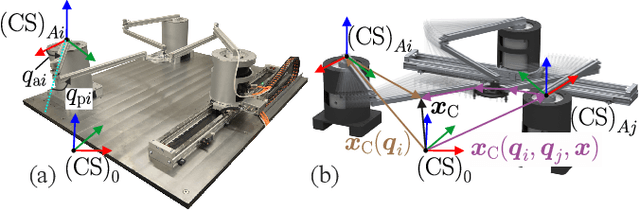
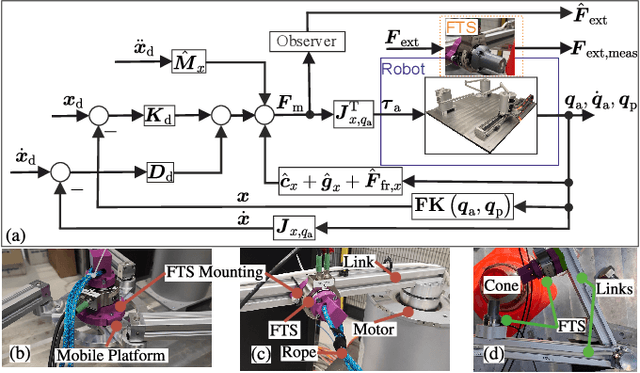
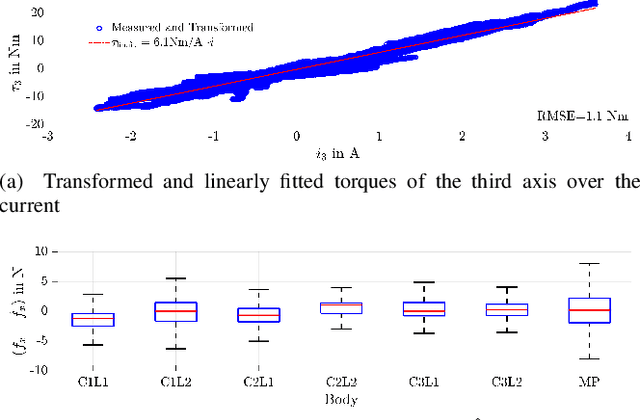
Abstract:Parallel robots provide the potential to be leveraged for human-robot collaboration (HRC) due to low collision energies even at high speeds resulting from their reduced moving masses. However, the risk of unintended contact with the leg chains increases compared to the structure of serial robots. As a first step towards HRC, contact cases on the whole parallel robot structure are investigated and a disturbance observer based on generalized momenta and measurements of motor current is applied. In addition, a Kalman filter and a second-order sliding-mode observer based on generalized momenta are compared in terms of error and detection time. Gearless direct drives with low friction improve external force estimation and enable low impedance. The experimental validation is performed with two force-torque sensors and a kinetostatic model. This allows a new identification method of the motor torque constant of an assembled parallel robot to estimate external forces from the motor current and via a dynamics model. A Cartesian impedance control scheme for compliant robot-environmental dynamics with stiffness from 0.1-2N/mm and the force observation for low forces over the entire structure are validated. The observers are used for collisions and clamping at velocities of 0.4-0.9m/s for detection within 9-58ms and a reaction in the form of a zero-g mode.
Collision Isolation and Identification Using Proprioceptive Sensing for Parallel Robots to Enable Human-Robot Collaboration
Aug 18, 2023Abstract:Parallel robots (PRs) allow for higher speeds in human-robot collaboration due to their lower moving masses but are more prone to unintended contact. For a safe reaction, knowledge of the location and force of a collision is useful. A novel algorithm for collision isolation and identification with proprioceptive information for a real PR is the scope of this work. To classify the collided body, the effects of contact forces at the links and platform of the PR are analyzed using a kinetostatic projection. This insight enables the derivation of features from the line of action of the estimated external force. The significance of these features is confirmed in experiments for various load cases. A feedforward neural network (FNN) classifies the collided body based on these physically modeled features. Generalization with the FNN to 300k load cases on the whole robot structure in other joint angle configurations is successfully performed with a collision-body classification accuracy of 84% in the experiments. Platform collisions are isolated and identified with an explicit solution, while a particle filter estimates the location and force of a contact on a kinematic chain. Updating the particle filter with estimated external joint torques leads to an isolation error of less than 3cm and an identification error of 4N in a real-world experiment.
Recalibration of Aleatoric and Epistemic Regression Uncertainty in Medical Imaging
Apr 26, 2021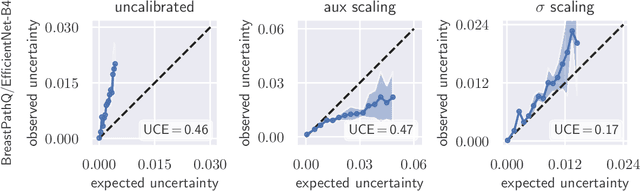
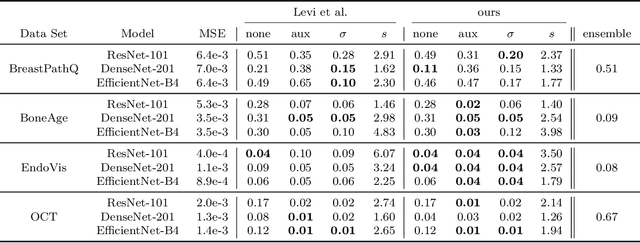
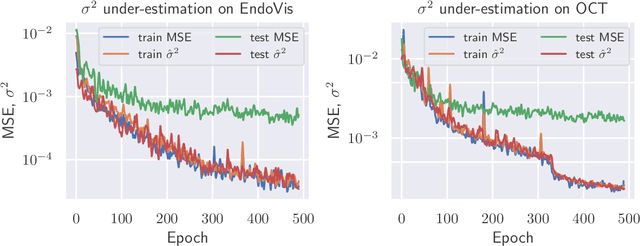
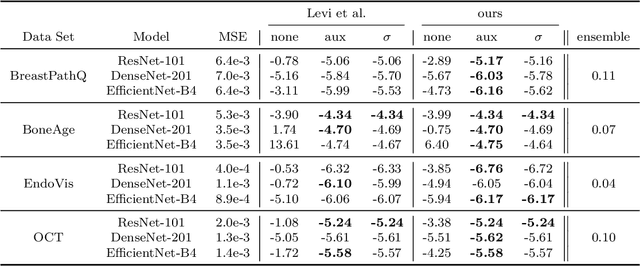
Abstract:The consideration of predictive uncertainty in medical imaging with deep learning is of utmost importance. We apply estimation of both aleatoric and epistemic uncertainty by variational Bayesian inference with Monte Carlo dropout to regression tasks and show that predictive uncertainty is systematically underestimated. We apply $ \sigma $ scaling with a single scalar value; a simple, yet effective calibration method for both types of uncertainty. The performance of our approach is evaluated on a variety of common medical regression data sets using different state-of-the-art convolutional network architectures. In our experiments, $ \sigma $ scaling is able to reliably recalibrate predictive uncertainty. It is easy to implement and maintains the accuracy. Well-calibrated uncertainty in regression allows robust rejection of unreliable predictions or detection of out-of-distribution samples. Our source code is available at https://github.com/mlaves/well-calibrated-regression-uncertainty
Uncertainty Estimation in Medical Image Denoising with Bayesian Deep Image Prior
Aug 20, 2020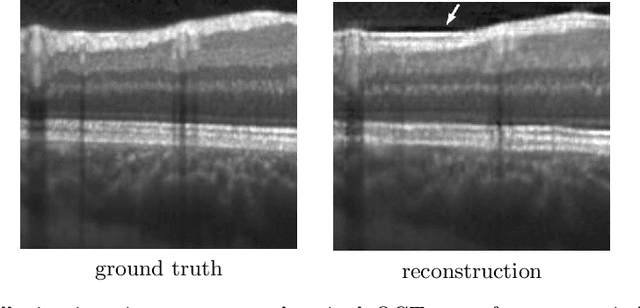
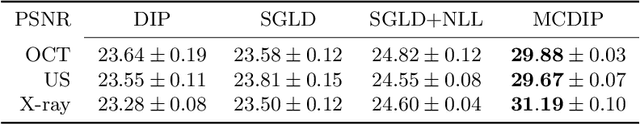
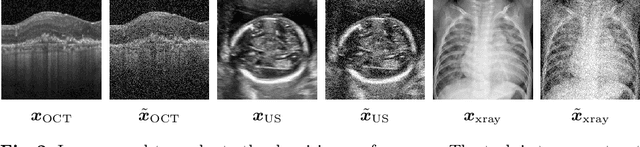

Abstract:Uncertainty quantification in inverse medical imaging tasks with deep learning has received little attention. However, deep models trained on large data sets tend to hallucinate and create artifacts in the reconstructed output that are not anatomically present. We use a randomly initialized convolutional network as parameterization of the reconstructed image and perform gradient descent to match the observation, which is known as deep image prior. In this case, the reconstruction does not suffer from hallucinations as no prior training is performed. We extend this to a Bayesian approach with Monte Carlo dropout to quantify both aleatoric and epistemic uncertainty. The presented method is evaluated on the task of denoising different medical imaging modalities. The experimental results show that our approach yields well-calibrated uncertainty. That is, the predictive uncertainty correlates with the predictive error. This allows for reliable uncertainty estimates and can tackle the problem of hallucinations and artifacts in inverse medical imaging tasks.
Patient-Specific Domain Adaptation for Fast Optical Flow Based on Teacher-Student Knowledge Transfer
Jul 09, 2020
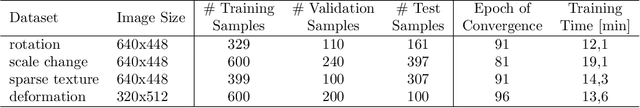
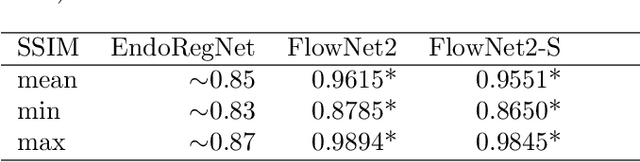
Abstract:Fast motion feedback is crucial in computer-aided surgery (CAS) on moving tissue. Image-assistance in safety-critical vision applications requires a dense tracking of tissue motion. This can be done using optical flow (OF). Accurate motion predictions at high processing rates lead to higher patient safety. Current deep learning OF models show the common speed vs. accuracy trade-off. To achieve high accuracy at high processing rates, we propose patient-specific fine-tuning of a fast model. This minimizes the domain gap between training and application data, while reducing the target domain to the capability of the lower complex, fast model. We propose to obtain training sequences pre-operatively in the operation room. We handle missing ground truth, by employing teacher-student learning. Using flow estimations from teacher model FlowNet2 we specialize a fast student model FlowNet2S on the patient-specific domain. Evaluation is performed on sequences from the Hamlyn dataset. Our student model shows very good performance after fine-tuning. Tracking accuracy is comparable to the teacher model at a speed up of factor six. Fine-tuning can be performed within minutes, making it feasible for the operation room. Our method allows to use a real-time capable model that was previously not suited for this task. This method is laying the path for improved patient-specific motion estimation in CAS.
Calibration of Model Uncertainty for Dropout Variational Inference
Jun 20, 2020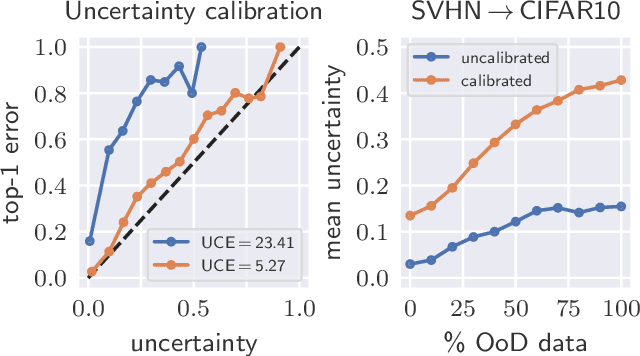
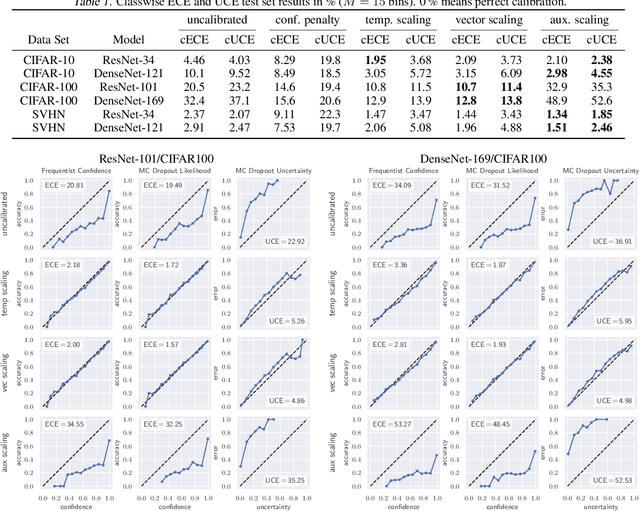
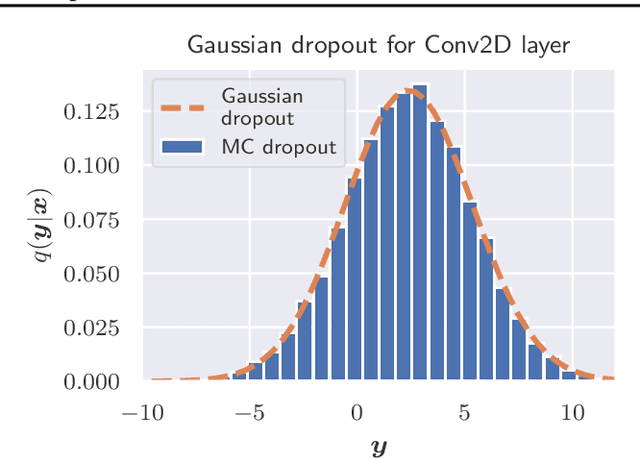
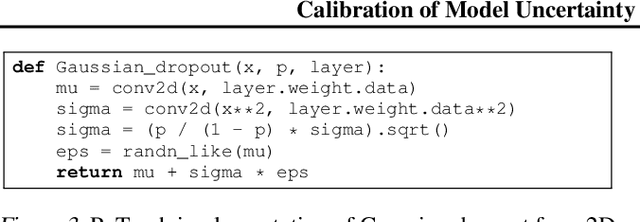
Abstract:The model uncertainty obtained by variational Bayesian inference with Monte Carlo dropout is prone to miscalibration. In this paper, different logit scaling methods are extended to dropout variational inference to recalibrate model uncertainty. Expected uncertainty calibration error (UCE) is presented as a metric to measure miscalibration. The effectiveness of recalibration is evaluated on CIFAR-10/100 and SVHN for recent CNN architectures. Experimental results show that logit scaling considerably reduce miscalibration by means of UCE. Well-calibrated uncertainty enables reliable rejection of uncertain predictions and robust detection of out-of-distribution data.
Well-calibrated Model Uncertainty with Temperature Scaling for Dropout Variational Inference
Oct 01, 2019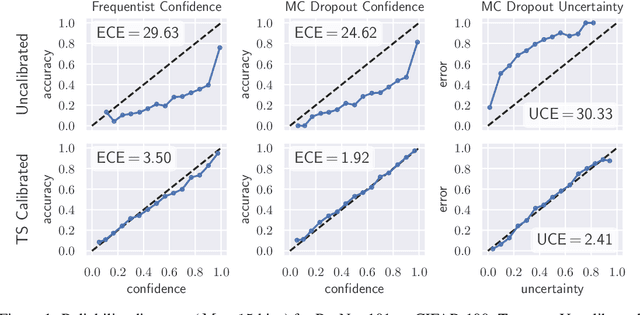
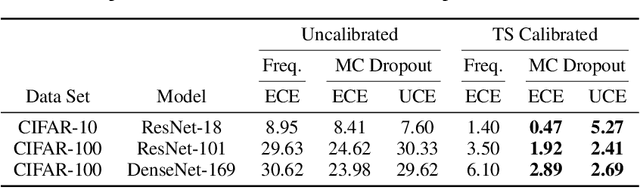
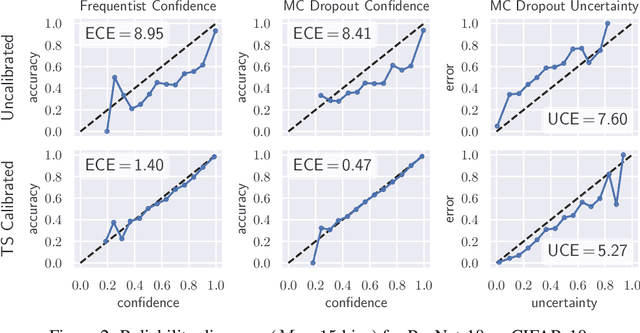
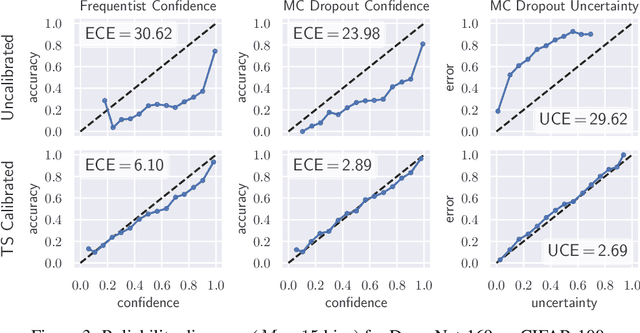
Abstract:In this paper, well-calibrated model uncertainty is obtained by using temperature scaling together with Monte Carlo dropout as approximation to Bayesian inference. The proposed approach can easily be derived from frequentist temperature scaling and yields well-calibrated model uncertainty as well as softmax likelihood.
Uncertainty Quantification in Computer-Aided Diagnosis: Make Your Model say "I don't know" for Ambiguous Cases
Aug 02, 2019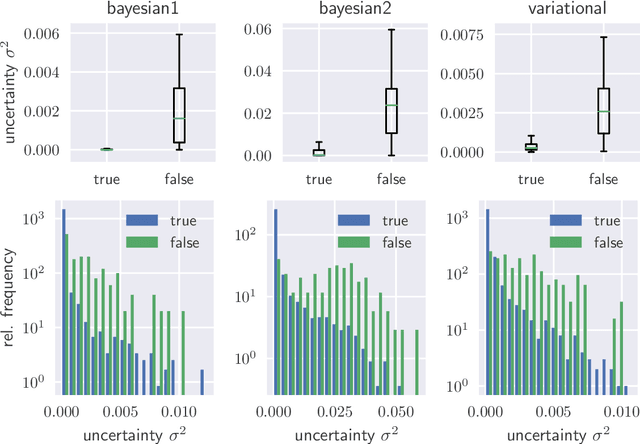

Abstract:We evaluate two different methods for the integration of prediction uncertainty into diagnostic image classifiers to increase patient safety in deep learning. In the first method, Monte Carlo sampling is applied with dropout at test time to get a posterior distribution of the class labels (Bayesian ResNet). The second method extends ResNet to a probabilistic approach by predicting the parameters of the posterior distribution and sampling the final result from it (Variational ResNet).The variance of the posterior is used as metric for uncertainty.Both methods are trained on a data set of optical coherence tomography scans showing four different retinal conditions. Our results shown that cases in which the classifier predicts incorrectly correlate with a higher uncertainty. Mean uncertainty of incorrectly diagnosed cases was between 4.6 and 8.1 times higher than mean uncertainty of correctly diagnosed cases. Modeling of the prediction uncertainty in computer-aided diagnosis with deep learning yields more reliable results and is anticipated to increase patient safety.
Deformable Medical Image Registration Using a Randomly-Initialized CNN as Regularization Prior
Aug 02, 2019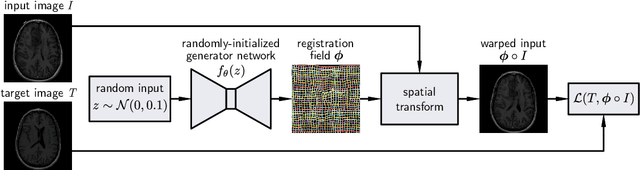
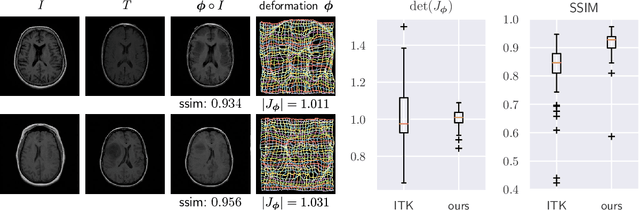
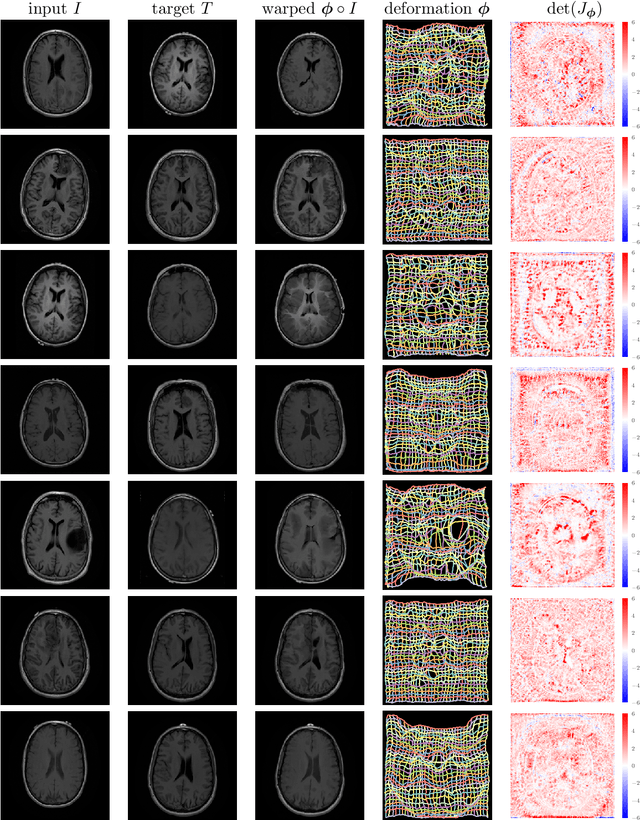
Abstract:We present deformable unsupervised medical image registration using a randomly-initialized deep convolutional neural network (CNN) as regularization prior. Conventional registration methods predict a transformation by minimizing dissimilarities between an image pair. The minimization is usually regularized with manually engineered priors, which limits the potential of the registration. By learning transformation priors from a large dataset, CNNs have achieved great success in deformable registration. However, learned methods are restricted to domain-specific data and the required amounts of medical data are difficult to obtain. Our approach uses the idea of deep image priors to combine convolutional networks with conventional registration methods based on manually engineered priors. The proposed method is applied to brain MRI scans. We show that our approach registers image pairs with state-of-the-art accuracy by providing dense, pixel-wise correspondence maps. It does not rely on prior training and is therefore not limited to a specific image domain.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge