Lüder A. Kahrs
Recalibration of Aleatoric and Epistemic Regression Uncertainty in Medical Imaging
Apr 26, 2021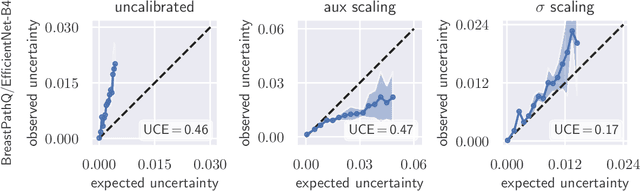
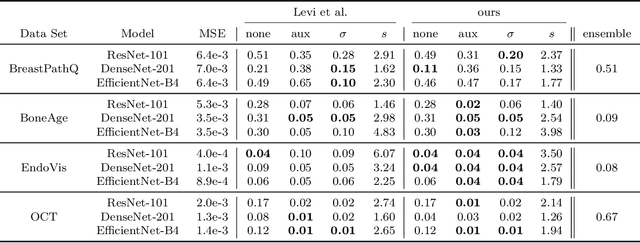
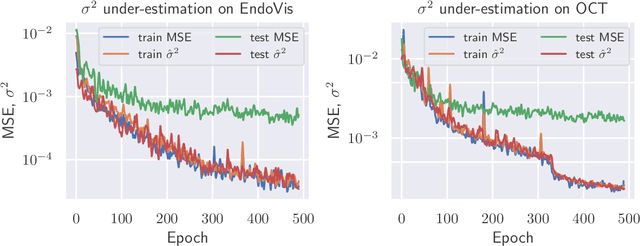
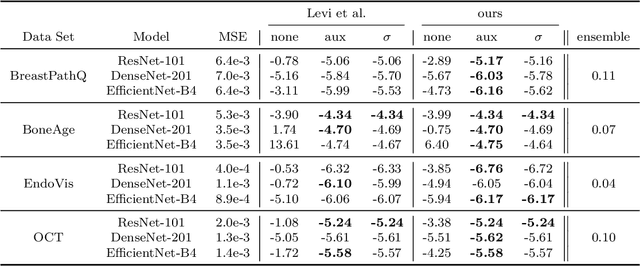
Abstract:The consideration of predictive uncertainty in medical imaging with deep learning is of utmost importance. We apply estimation of both aleatoric and epistemic uncertainty by variational Bayesian inference with Monte Carlo dropout to regression tasks and show that predictive uncertainty is systematically underestimated. We apply $ \sigma $ scaling with a single scalar value; a simple, yet effective calibration method for both types of uncertainty. The performance of our approach is evaluated on a variety of common medical regression data sets using different state-of-the-art convolutional network architectures. In our experiments, $ \sigma $ scaling is able to reliably recalibrate predictive uncertainty. It is easy to implement and maintains the accuracy. Well-calibrated uncertainty in regression allows robust rejection of unreliable predictions or detection of out-of-distribution samples. Our source code is available at https://github.com/mlaves/well-calibrated-regression-uncertainty
Retinal OCT disease classification with variational autoencoder regularization
Mar 23, 2019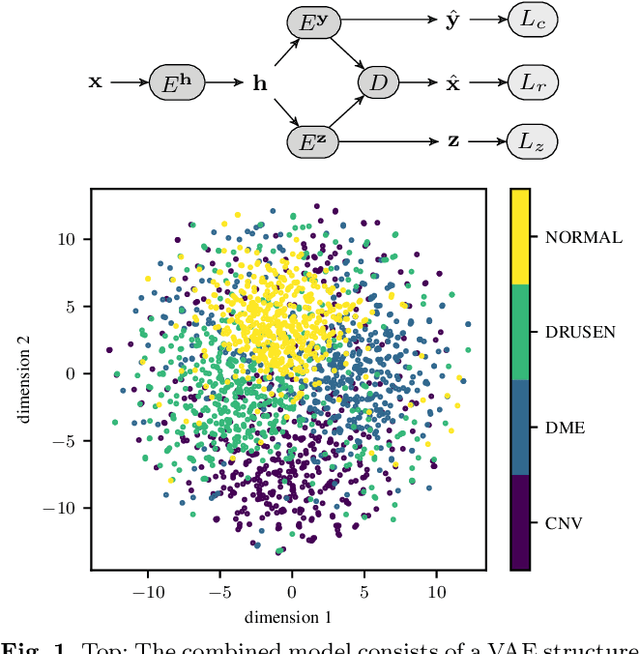
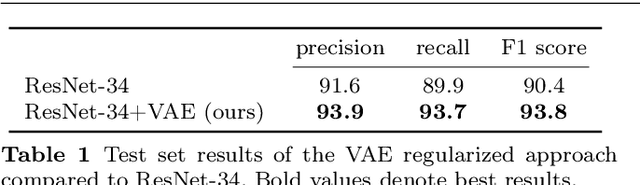
Abstract:According to the World Health Organization, 285 million people worldwide live with visual impairment. The most commonly used imaging technique for diagnosis in ophthalmology is optical coherence tomography (OCT). However, analysis of retinal OCT requires trained ophthalmologists and time, making a comprehensive early diagnosis unlikely. A recent study established a diagnostic tool based on convolutional neural networks (CNN), which was trained on a large database of retinal OCT images. The performance of the tool in classifying retinal conditions was on par to that of trained medical experts. However, the training of these networks is based on an enormous amount of labeled data, which is expensive and difficult to obtain. Therefore, this paper describes a method based on variational autoencoder regularization that improves classification performance when using a limited amount of labeled data. This work uses a two-path CNN model combining a classification network with an autoencoder (AE) for regularization. The key idea behind this is to prevent overfitting when using a limited training dataset size with small number of patients. Results show superior classification performance compared to a pre-trained and fully fine-tuned baseline ResNet-34. Clustering of the latent space in relation to the disease class is distinct. Neural networks for disease classification on OCTs can benefit from regularization using variational autoencoders when trained with limited amount of patient data. Especially in the medical imaging domain, data annotated by experts is expensive to obtain.
Endoscopic vs. volumetric OCT imaging of mastoid bone structure for pose estimation in minimally invasive cochlear implant surgery
Mar 23, 2019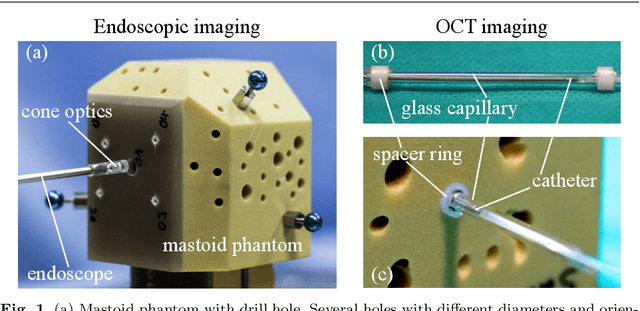
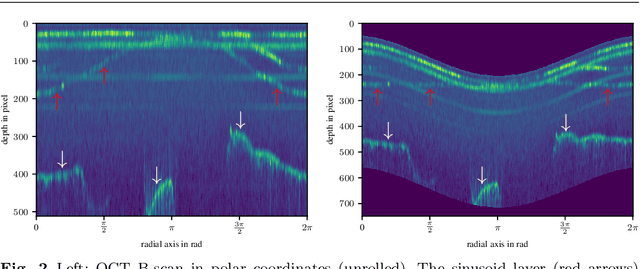
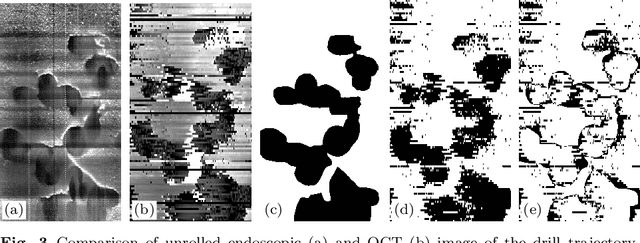
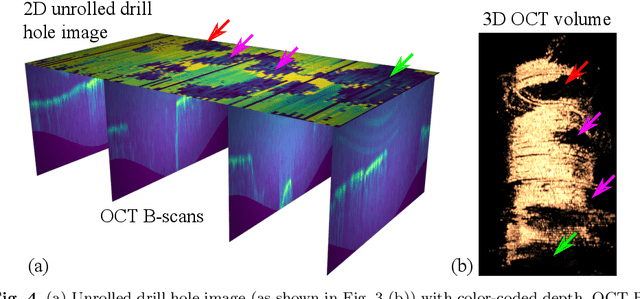
Abstract:Purpose: The facial recess is a delicate structure that must be protected in minimally invasive cochlear implant surgery. Current research estimates the drill trajectory by using endoscopy of the unique mastoid patterns. However, missing depth information limits available features for a registration to preoperative CT data. Therefore, this paper evaluates OCT for enhanced imaging of drill holes in mastoid bone and compares OCT data to original endoscopic images. Methods: A catheter-based OCT probe is inserted into a drill trajectory of a mastoid phantom in a translation-rotation manner to acquire the inner surface state. The images are undistorted and stitched to create volumentric data of the drill hole. The mastoid cell pattern is segmented automatically and compared to ground truth. Results: The mastoid pattern segmented on images acquired with OCT show a similarity of J = 73.6 % to ground truth based on endoscopic images and measured with the Jaccard metric. Leveraged by additional depth information, automated segmentation tends to be more robust and fail-safe compared to endoscopic images. Conclusion: The feasibility of using a clinically approved OCT probe for imaging the drill hole in cochlear implantation is shown. The resulting volumentric images provide additional information on the shape of caveties in the bone structure, which will be useful for image-to-patient registration and to estimate the drill trajectory. This will be another step towards safe minimally invasive cochlear implantation.
Deep learning based 2.5D flow field estimation for maximum intensity projections of 4D optical coherence tomography
Oct 26, 2018Abstract:In laser microsurgery, image-based control of the ablation laser can lead to higher accuracy and patient safety. However, camera-based image acquisition lacks the subcutaneous tissue perception. Optical coherence tomography (OCT) as high-resolution imaging modality yields transsectional images of tissue and can provide the missing depth information. Therefore, this paper deals with the tracking of distinctive subcutaneous structures on OCTs for automated control of ablation lasers in microsurgery. We present a deep learning based tracking scheme for concise representations of subsequent 3D OCT volumes. For each of the volume, a compact representation is created by calculating maximum intensity projections and projecting the depth value, were the maximum intensity voxel is found, onto an image plane. These depth images are then used for tracking by estimating the dense optical flow and depth changes with a self-supervisely trained convolutional neural network. Tracking performances are evaluated on a dataset of ex vivo human temporal bone with rigid ground truth transformations and on an in vivo sequence of human skin with non-rigid transformations. First quantitative evaluation reveals a mean endpoint error of 2.27voxel for scene flow estimation. Object tracking on 4D OCT data enables its use for sub-epithelial tracking of tissue structures for image-guidance in automated laser incision control for microsurgery.
A Dataset of Laryngeal Endoscopic Images with Comparative Study on Convolution Neural Network Based Semantic Segmentation
Sep 18, 2018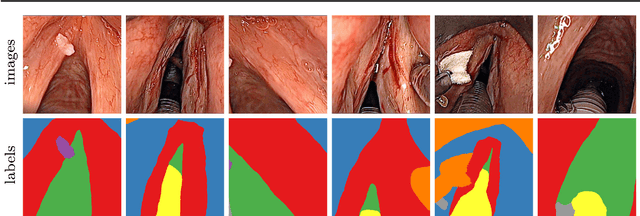



Abstract:Purpose: Automated segmentation of anatomical structures in medical image analysis is a key step in defining topology to enable or assist autonomous intervention robots. Recent methods based on deep convolutional neural networks (CNN) have outperformed former heuristic methods. However, those methods were primarily evaluated on rigid, real-world environments. In this study, we evaluate existing segmentation methods for their use with soft tissue. Methods: The four CNN-based methods SegNet, UNet, ENet and ErfNet are trained with high supervision on a novel 7-class dataset of surgeries on the human larynx. The dataset contains 400 manually segmented images from two patients during laser incisions. The Intersection-over-Union (IoU) evaluation metric is used to measure the accuracy of each method. Data augmentation and network ensembling is employed to increase segmentation accuracy. Stochastic inference is used to show the uncertainty of the individual models. Results: Our study shows that an average ensemble network of UNet and ErfNet is best suited for laryngeal soft tissue segmentation with a mean IoU of 84.7 %. The highest efficiency is achieved by ENet with a mean inference time of 9.22 ms per image on an NVIDIA GeForce GTX 1080 Ti GPU. All methods can be improved by data augmentation. Conclusion: CNN-based methods for semantic segmentation are applicable to laryngeal soft tissue. The segmentation can be used for active constraints or autonomous control in robot-assisted laser surgery. Further improvements could be achieved by using a larger dataset or training the models in a self-supervised manner on additional unlabeled data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge