Sarah Latus
Automated Estimation of Anatomical Risk Metrics for Endoscopic Sinus Surgery Using Deep Learning
Nov 10, 2025Abstract:Endoscopic sinus surgery requires careful preoperative assessment of the skull base anatomy to minimize risks such as cerebrospinal fluid leakage. Anatomical risk scores like the Keros, Gera and Thailand-Malaysia-Singapore score offer a standardized approach but require time-consuming manual measurements on coronal CT or CBCT scans. We propose an automated deep learning pipeline that estimates these risk scores by localizing key anatomical landmarks via heatmap regression. We compare a direct approach to a specialized global-to-local learning strategy and find mean absolute errors on the relevant anatomical measurements of 0.506mm for the Keros, 4.516° for the Gera and 0.802mm / 0.777mm for the TMS classification.
Tracking Any Point Methods for Markerless 3D Tissue Tracking in Endoscopic Stereo Images
Aug 11, 2025Abstract:Minimally invasive surgery presents challenges such as dynamic tissue motion and a limited field of view. Accurate tissue tracking has the potential to support surgical guidance, improve safety by helping avoid damage to sensitive structures, and enable context-aware robotic assistance during complex procedures. In this work, we propose a novel method for markerless 3D tissue tracking by leveraging 2D Tracking Any Point (TAP) networks. Our method combines two CoTracker models, one for temporal tracking and one for stereo matching, to estimate 3D motion from stereo endoscopic images. We evaluate the system using a clinical laparoscopic setup and a robotic arm simulating tissue motion, with experiments conducted on a synthetic 3D-printed phantom and a chicken tissue phantom. Tracking on the chicken tissue phantom yielded more reliable results, with Euclidean distance errors as low as 1.1 mm at a velocity of 10 mm/s. These findings highlight the potential of TAP-based models for accurate, markerless 3D tracking in challenging surgical scenarios.
A Mobile Robotic Approach to Autonomous Surface Scanning in Legal Medicine
Feb 20, 2025Abstract:Purpose: Comprehensive legal medicine documentation includes both an internal but also an external examination of the corpse. Typically, this documentation is conducted manually during conventional autopsy. A systematic digital documentation would be desirable, especially for the external examination of wounds, which is becoming more relevant for legal medicine analysis. For this purpose, RGB surface scanning has been introduced. While a manual full surface scan using a handheld camera is timeconsuming and operator dependent, floor or ceiling mounted robotic systems require substantial space and a dedicated room. Hence, we consider whether a mobile robotic system can be used for external documentation. Methods: We develop a mobile robotic system that enables full-body RGB-D surface scanning. Our work includes a detailed configuration space analysis to identify the environmental parameters that need to be considered to successfully perform a surface scan. We validate our findings through an experimental study in the lab and demonstrate the system's application in a legal medicine environment. Results: Our configuration space analysis shows that a good trade-off between coverage and time is reached with three robot base positions, leading to a coverage of 94.96 %. Experiments validate the effectiveness of the system in accurately capturing body surface geometry with an average surface coverage of 96.90 +- 3.16 % and 92.45 +- 1.43 % for a body phantom and actual corpses, respectively. Conclusion: This work demonstrates the potential of a mobile robotic system to automate RGB-D surface scanning in legal medicine, complementing the use of post-mortem CT scans for inner documentation. Our results indicate that the proposed system can contribute to more efficient and autonomous legal medicine documentation, reducing the need for manual intervention.
A Modified da Vinci Surgical Instrument for OCE based Elasticity Estimation with Deep Learning
Mar 14, 2024



Abstract:Robot-assisted surgery has advantages compared to conventional laparoscopic procedures, e.g., precise movement of the surgical instruments, improved dexterity, and high-resolution visualization of the surgical field. However, mechanical tissue properties may provide additional information, e.g., on the location of lesions or vessels. While elastographic imaging has been proposed, it is not readily available as an online modality during robot-assisted surgery. We propose modifying a da~Vinci surgical instrument to realize optical coherence elastography (OCE) for quantitative elasticity estimation. The modified da~Vinci instrument is equipped with piezoelectric elements for shear wave excitation and we employ fast optical coherence tomography (OCT) imaging to track propagating wave fields, which are directly related to biomechanical tissue properties. All high-voltage components are mounted at the proximal end outside the patient. We demonstrate that external excitation at the instrument shaft can effectively stimulate shear waves, even when considering damping. Comparing conventional and deep learning-based signal processing, resulting in mean absolute errors of 19.27 kPa and 6.29 kPa, respectively. These results illustrate that precise quantitative elasticity estimates can be obtained. We also demonstrate quantitative elasticity estimation on ex-vivo tissue samples of heart, liver and stomach, and show that the measurements can be used to distinguish soft and stiff tissue types.
Collaborative Robotic Biopsy with Trajectory Guidance and Needle Tip Force Feedback
Jun 12, 2023Abstract:The diagnostic value of biopsies is highly dependent on the placement of needles. Robotic trajectory guidance has been shown to improve needle positioning, but feedback for real-time navigation is limited. Haptic display of needle tip forces can provide rich feedback for needle navigation by enabling localization of tissue structures along the insertion path. We present a collaborative robotic biopsy system that combines trajectory guidance with kinesthetic feedback to assist the physician in needle placement. The robot aligns the needle while the insertion is performed in collaboration with a medical expert who controls the needle position on site. We present a needle design that senses forces at the needle tip based on optical coherence tomography and machine learning for real-time data processing. Our robotic setup allows operators to sense deep tissue interfaces independent of frictional forces to improve needle placement relative to a desired target structure. We first evaluate needle tip force sensing in ex-vivo tissue in a phantom study. We characterize the tip forces during insertions with constant velocity and demonstrate the ability to detect tissue interfaces in a collaborative user study. Participants are able to detect 91% of ex-vivo tissue interfaces based on needle tip force feedback alone. Finally, we demonstrate that even smaller, deep target structures can be accurately sampled by performing post-mortem in situ biopsies of the pancreas.
Tissue Classification During Needle Insertion Using Self-Supervised Contrastive Learning and Optical Coherence Tomography
Apr 26, 2023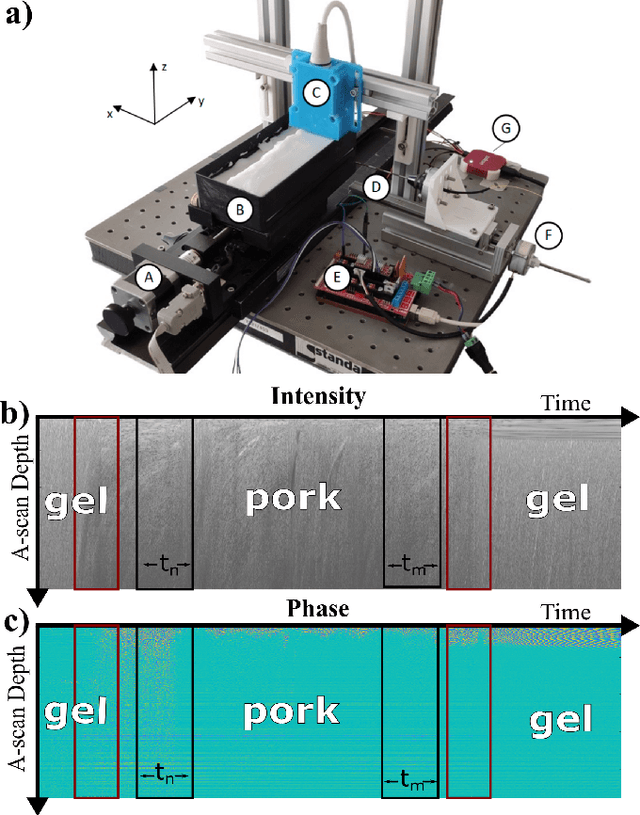
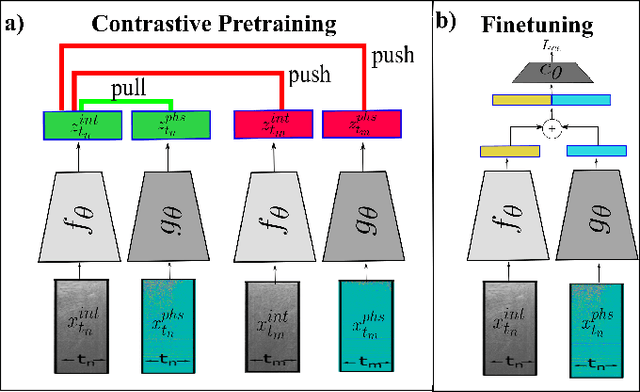
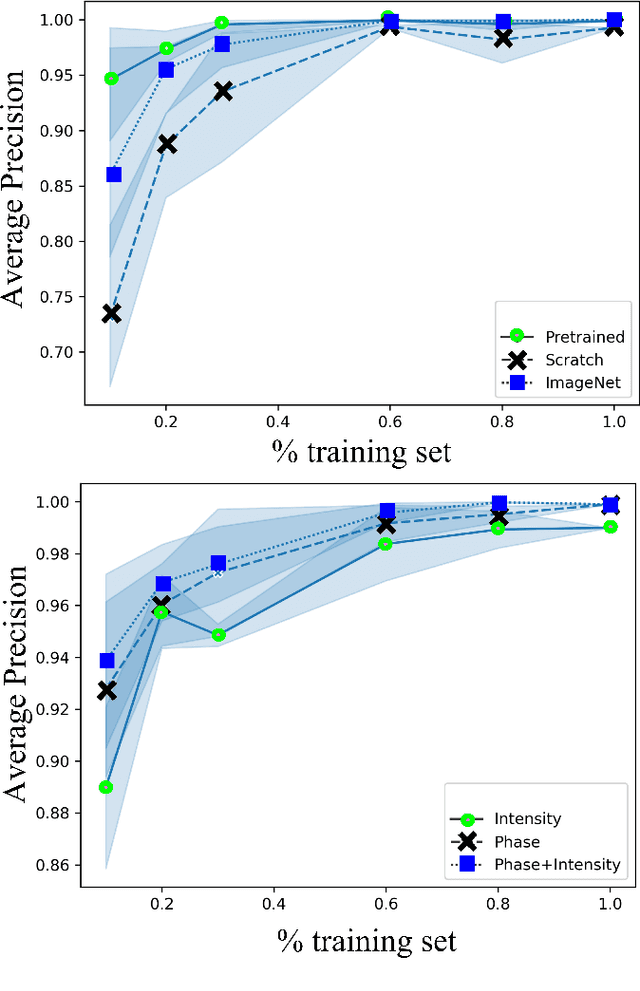
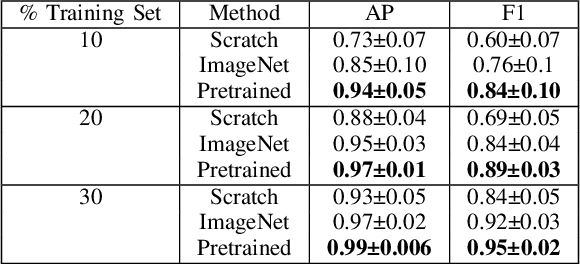
Abstract:Needle positioning is essential for various medical applications such as epidural anaesthesia. Physicians rely on their instincts while navigating the needle in epidural spaces. Thereby, identifying the tissue structures may be helpful to the physician as they can provide additional feedback in the needle insertion process. To this end, we propose a deep neural network that classifies the tissues from the phase and intensity data of complex OCT signals acquired at the needle tip. We investigate the performance of the deep neural network in a limited labelled dataset scenario and propose a novel contrastive pretraining strategy that learns invariant representation for phase and intensity data. We show that with 10% of the training set, our proposed pretraining strategy helps the model achieve an F1 score of 0.84 whereas the model achieves an F1 score of 0.60 without it. Further, we analyse the importance of phase and intensity individually towards tissue classification.
Ultrasound Shear Wave Elasticity Imaging with Spatio-Temporal Deep Learning
Apr 28, 2022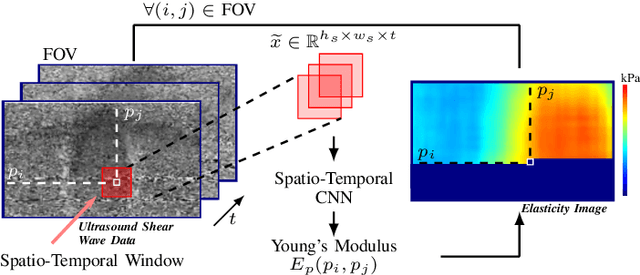
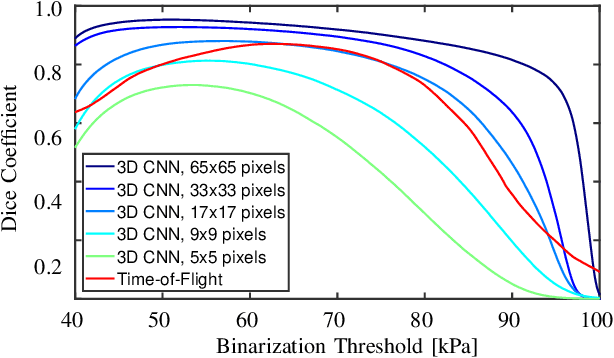
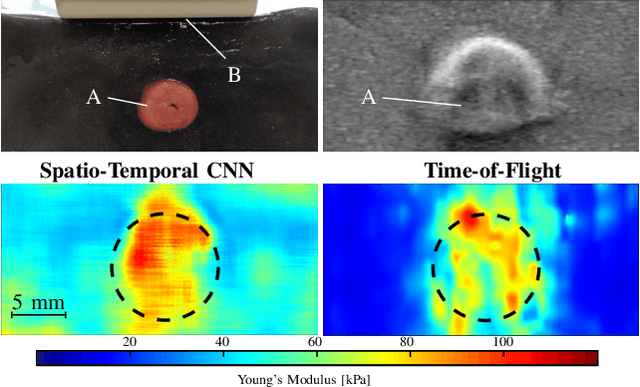
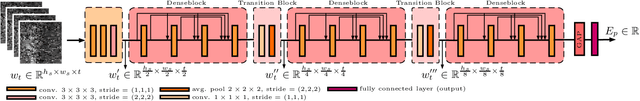
Abstract:Ultrasound shear wave elasticity imaging is a valuable tool for quantifying the elastic properties of tissue. Typically, the shear wave velocity is derived and mapped to an elasticity value, which neglects information such as the shape of the propagating shear wave or push sequence characteristics. We present 3D spatio-temporal CNNs for fast local elasticity estimation from ultrasound data. This approach is based on retrieving elastic properties from shear wave propagation within small local regions. A large training data set is acquired with a robot from homogeneous gelatin phantoms ranging from 17.42 kPa to 126.05 kPa with various push locations. The results show that our approach can estimate elastic properties on a pixelwise basis with a mean absolute error of 5.01+-4.37 kPa. Furthermore, we estimate local elasticity independent of the push location and can even perform accurate estimates inside the push region. For phantoms with embedded inclusions, we report a 53.93% lower MAE (7.50 kPa) and on the background of 85.24% (1.64 kPa) compared to a conventional shear wave method. Overall, our method offers fast local estimations of elastic properties with small spatio-temporal window sizes.
A novel optical needle probe for deep learning-based tissue elasticity characterization
Sep 20, 2021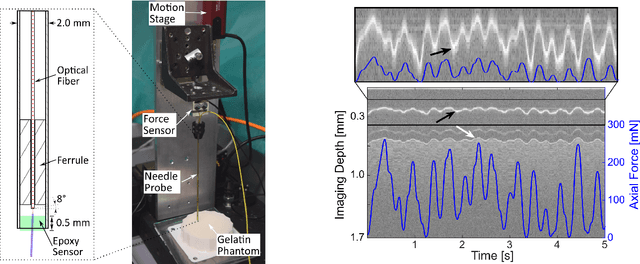
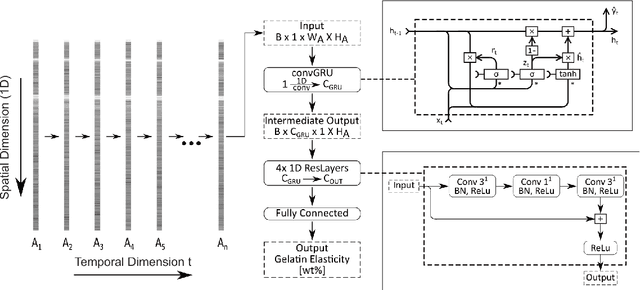
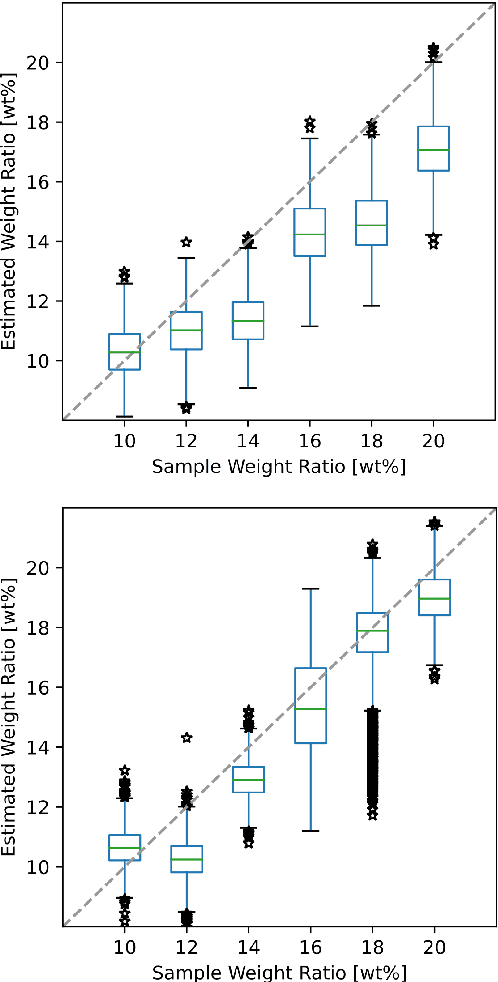
Abstract:The distinction between malignant and benign tumors is essential to the treatment of cancer. The tissue's elasticity can be used as an indicator for the required tissue characterization. Optical coherence elastography (OCE) probes have been proposed for needle insertions but have so far lacked the necessary load sensing capabilities. We present a novel OCE needle probe that provides simultaneous optical coherence tomography (OCT) imaging and load sensing at the needle tip. We demonstrate the application of the needle probe in indentation experiments on gelatin phantoms with varying gelatin concentrations. We further implement two deep learning methods for the end-to-end sample characterization from the acquired OCT data. We report the estimation of gelatin sample concentrations in unseen samples with a mean error of $1.21 \pm 0.91$ wt\%. Both evaluated deep learning models successfully provide sample characterization with different advantages regarding the accuracy and inference time.
* Accepted at CURAC 2021, 2nd Place in the Best Paper Awards
Endoscopic vs. volumetric OCT imaging of mastoid bone structure for pose estimation in minimally invasive cochlear implant surgery
Mar 23, 2019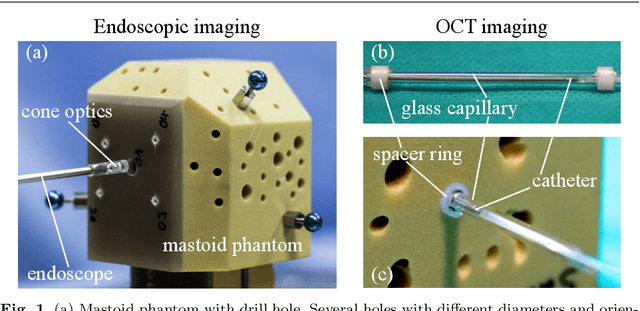
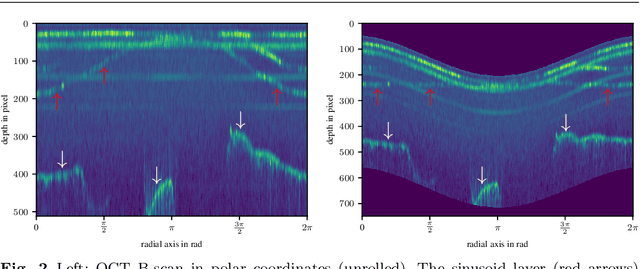
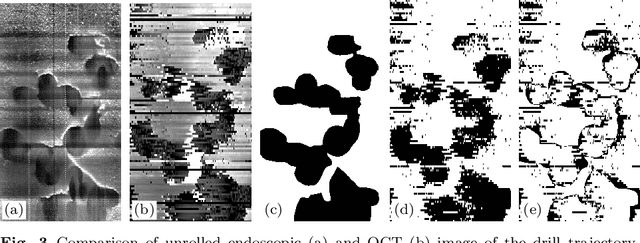
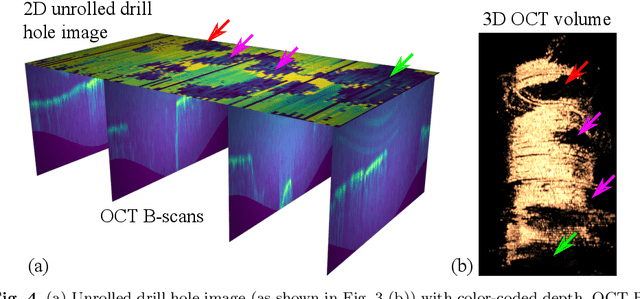
Abstract:Purpose: The facial recess is a delicate structure that must be protected in minimally invasive cochlear implant surgery. Current research estimates the drill trajectory by using endoscopy of the unique mastoid patterns. However, missing depth information limits available features for a registration to preoperative CT data. Therefore, this paper evaluates OCT for enhanced imaging of drill holes in mastoid bone and compares OCT data to original endoscopic images. Methods: A catheter-based OCT probe is inserted into a drill trajectory of a mastoid phantom in a translation-rotation manner to acquire the inner surface state. The images are undistorted and stitched to create volumentric data of the drill hole. The mastoid cell pattern is segmented automatically and compared to ground truth. Results: The mastoid pattern segmented on images acquired with OCT show a similarity of J = 73.6 % to ground truth based on endoscopic images and measured with the Jaccard metric. Leveraged by additional depth information, automated segmentation tends to be more robust and fail-safe compared to endoscopic images. Conclusion: The feasibility of using a clinically approved OCT probe for imaging the drill hole in cochlear implantation is shown. The resulting volumentric images provide additional information on the shape of caveties in the bone structure, which will be useful for image-to-patient registration and to estimate the drill trajectory. This will be another step towards safe minimally invasive cochlear implantation.
Towards Automatic Lesion Classification in the Upper Aerodigestive Tract Using OCT and Deep Transfer Learning Methods
Feb 10, 2019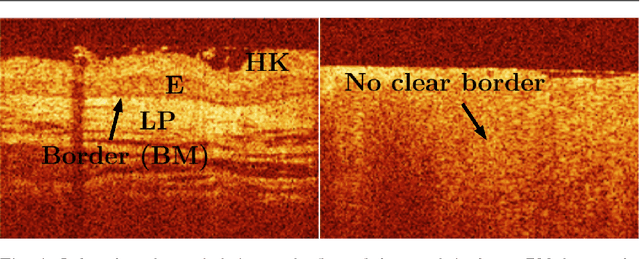
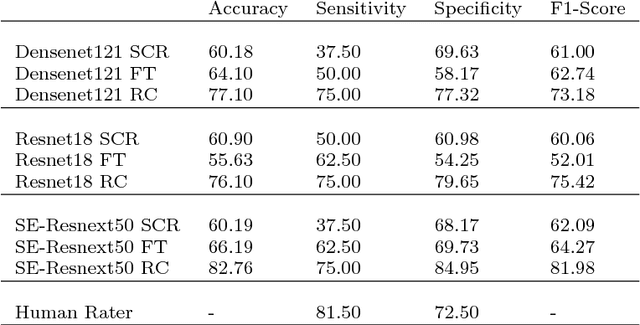
Abstract:Early detection of cancer is crucial for treatment and overall patient survival. In the upper aerodigestive tract (UADT) the gold standard for identification of malignant tissue is an invasive biopsy. Recently, non-invasive imaging techniques such as confocal laser microscopy and optical coherence tomography (OCT) have been used for tissue assessment. In particular, in a recent study experts classified lesions in the UADT with respect to their invasiveness using OCT images only. As the results were promising, automatic classification of lesions might be feasible which could assist experts in their decision making. Therefore, we address the problem of automatic lesion classification from OCT images. This task is very challenging as the available dataset is extremely small and the data quality is limited. However, as similar issues are typical in many clinical scenarios we study to what extent deep learning approaches can still be trained and used for decision support.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge