David M. Leistner
Bioresorbable Scaffold Visualization in IVOCT Images Using CNNs and Weakly Supervised Localization
Oct 22, 2018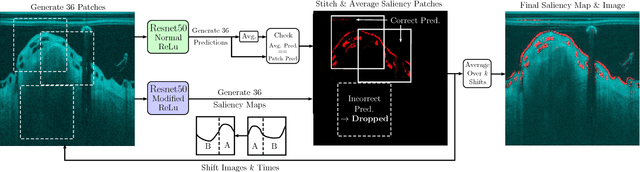
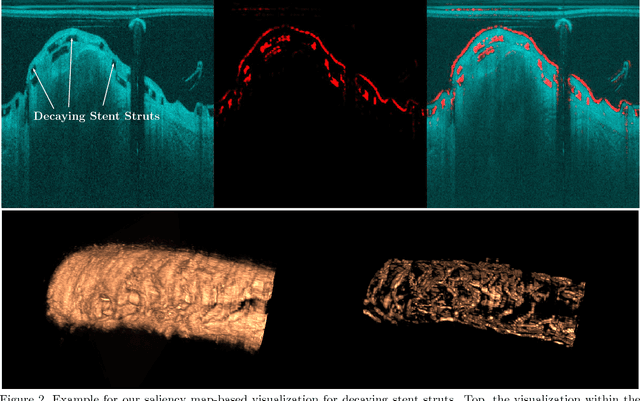
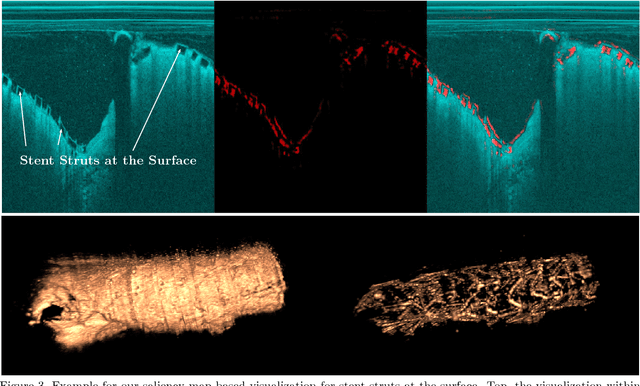
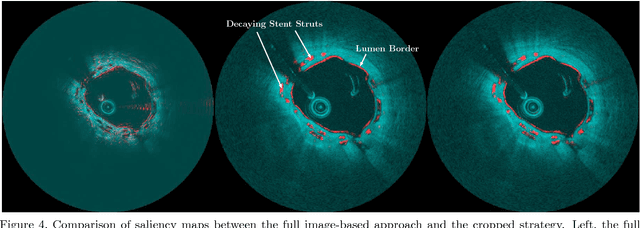
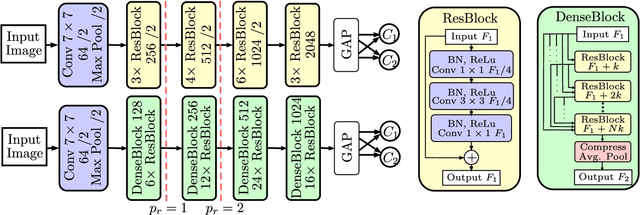
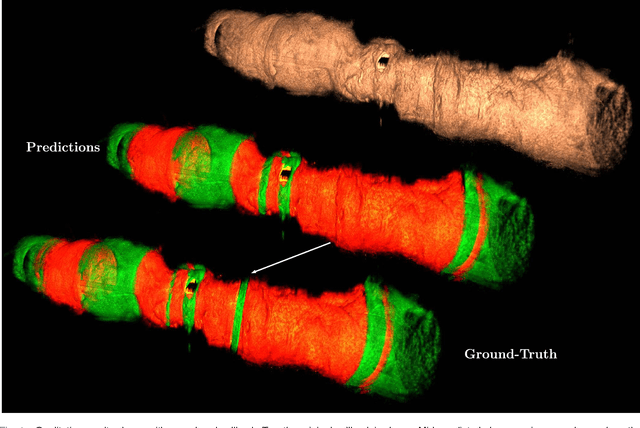
Abstract:Bioresorbable scaffolds have become a popular choice for treatment of coronary heart disease, replacing traditional metal stents. Often, intravascular optical coherence tomography is used to assess potential malapposition after implantation and for follow-up examinations later on. Typically, the scaffold is manually reviewed by an expert, analyzing each of the hundreds of image slices. As this is time consuming, automatic stent detection and visualization approaches have been proposed, mostly for metal stent detection based on classic image processing. As bioresorbable scaffolds are harder to detect, recent approaches have used feature extraction and machine learning methods for automatic detection. However, these methods require detailed, pixel-level labels in each image slice and extensive feature engineering for the particular stent type which might limit the approaches' generalization capabilities. Therefore, we propose a deep learning-based method for bioresorbable scaffold visualization using only image-level labels. A convolutional neural network is trained to predict whether an image slice contains a metal stent, a bioresorbable scaffold, or no device. Then, we derive local stent strut information by employing weakly supervised localization using saliency maps with guided backpropagation. As saliency maps are generally diffuse and noisy, we propose a novel patch-based method with image shifting which allows for high resolution stent visualization. Our convolutional neural network model achieves a classification accuracy of 99.0 % for image-level stent classification which can be used for both high quality in-slice stent visualization and 3D rendering of the stent structure.
Automatic Plaque Detection in IVOCT Pullbacks Using Convolutional Neural Networks
Aug 24, 2018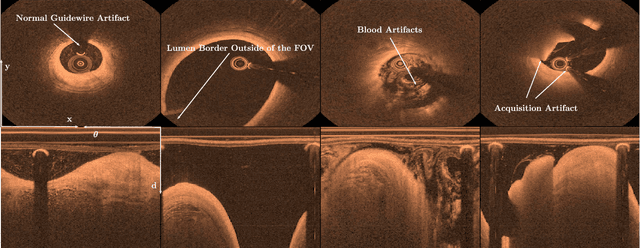

Abstract:Coronary heart disease is a common cause of death despite being preventable. To treat the underlying plaque deposits in the arterial walls, intravascular optical coherence tomography can be used by experts to detect and characterize the lesions. In clinical routine, hundreds of images are acquired for each patient which requires automatic plaque detection for fast and accurate decision support. So far, automatic approaches rely on classic machine learning methods and deep learning solutions have rarely been studied. Given the success of deep learning methods with other imaging modalities, a thorough understanding of deep learning-based plaque detection for future clinical decision support systems is required. We address this issue with a new dataset consisting of in-vivo patient images labeled by three trained experts. Using this dataset, we employ state-of-the-art deep learning models that directly learn plaque classification from the images. For improved performance, we study different transfer learning approaches. Furthermore, we investigate the use of cartesian and polar image representations and employ data augmentation techniques tailored to each representation. We fuse both representations in a multi-path architecture for more effective feature exploitation. Last, we address the challenge of plaque differentiation in addition to detection. Overall, we find that our combined model performs best with an accuracy of 91.7%, a sensitivity of 90.9% and a specificity of 92.4%. Our results indicate that building a deep learning-based clinical decision support system for plaque detection is feasible.
Adversarial Training for Patient-Independent Feature Learning with IVOCT Data for Plaque Classification
May 16, 2018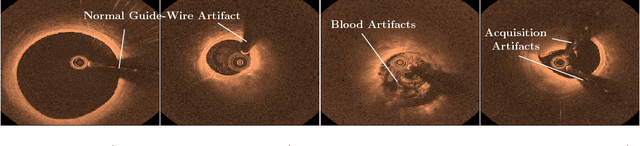

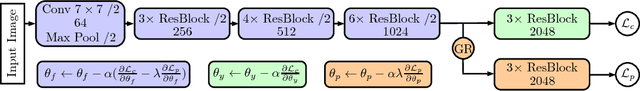
Abstract:Deep learning methods have shown impressive results for a variety of medical problems over the last few years. However, datasets tend to be small due to time-consuming annotation. As datasets with different patients are often very heterogeneous generalization to new patients can be difficult. This is complicated further if large differences in image acquisition can occur, which is common during intravascular optical coherence tomography for coronary plaque imaging. We address this problem with an adversarial training strategy where we force a part of a deep neural network to learn features that are independent of patient- or acquisitionspecific characteristics. We compare our regularization method to typical data augmentation strategies and show that our approach improves performance for a small medical dataset.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge