Robin Mieling
Leveraging the Mahalanobis Distance to enhance Unsupervised Brain MRI Anomaly Detection
Jul 17, 2024

Abstract:Unsupervised Anomaly Detection (UAD) methods rely on healthy data distributions to identify anomalies as outliers. In brain MRI, a common approach is reconstruction-based UAD, where generative models reconstruct healthy brain MRIs, and anomalies are detected as deviations between input and reconstruction. However, this method is sensitive to imperfect reconstructions, leading to false positives that impede the segmentation. To address this limitation, we construct multiple reconstructions with probabilistic diffusion models. We then analyze the resulting distribution of these reconstructions using the Mahalanobis distance to identify anomalies as outliers. By leveraging information about normal variations and covariance of individual pixels within this distribution, we effectively refine anomaly scoring, leading to improved segmentation. Our experimental results demonstrate substantial performance improvements across various data sets. Specifically, compared to relying solely on single reconstructions, our approach achieves relative improvements of 15.9%, 35.4%, 48.0%, and 4.7% in terms of AUPRC for the BRATS21, ATLAS, MSLUB and WMH data sets, respectively.
Diffusion Models with Ensembled Structure-Based Anomaly Scoring for Unsupervised Anomaly Detection
Mar 21, 2024


Abstract:Supervised deep learning techniques show promise in medical image analysis. However, they require comprehensive annotated data sets, which poses challenges, particularly for rare diseases. Consequently, unsupervised anomaly detection (UAD) emerges as a viable alternative for pathology segmentation, as only healthy data is required for training. However, recent UAD anomaly scoring functions often focus on intensity only and neglect structural differences, which impedes the segmentation performance. This work investigates the potential of Structural Similarity (SSIM) to bridge this gap. SSIM captures both intensity and structural disparities and can be advantageous over the classical $l1$ error. However, we show that there is more than one optimal kernel size for the SSIM calculation for different pathologies. Therefore, we investigate an adaptive ensembling strategy for various kernel sizes to offer a more pathology-agnostic scoring mechanism. We demonstrate that this ensembling strategy can enhance the performance of DMs and mitigate the sensitivity to different kernel sizes across varying pathologies, highlighting its promise for brain MRI anomaly detection.
A Modified da Vinci Surgical Instrument for OCE based Elasticity Estimation with Deep Learning
Mar 14, 2024



Abstract:Robot-assisted surgery has advantages compared to conventional laparoscopic procedures, e.g., precise movement of the surgical instruments, improved dexterity, and high-resolution visualization of the surgical field. However, mechanical tissue properties may provide additional information, e.g., on the location of lesions or vessels. While elastographic imaging has been proposed, it is not readily available as an online modality during robot-assisted surgery. We propose modifying a da~Vinci surgical instrument to realize optical coherence elastography (OCE) for quantitative elasticity estimation. The modified da~Vinci instrument is equipped with piezoelectric elements for shear wave excitation and we employ fast optical coherence tomography (OCT) imaging to track propagating wave fields, which are directly related to biomechanical tissue properties. All high-voltage components are mounted at the proximal end outside the patient. We demonstrate that external excitation at the instrument shaft can effectively stimulate shear waves, even when considering damping. Comparing conventional and deep learning-based signal processing, resulting in mean absolute errors of 19.27 kPa and 6.29 kPa, respectively. These results illustrate that precise quantitative elasticity estimates can be obtained. We also demonstrate quantitative elasticity estimation on ex-vivo tissue samples of heart, liver and stomach, and show that the measurements can be used to distinguish soft and stiff tissue types.
Guided Reconstruction with Conditioned Diffusion Models for Unsupervised Anomaly Detection in Brain MRIs
Dec 07, 2023



Abstract:Unsupervised anomaly detection in Brain MRIs aims to identify abnormalities as outliers from a healthy training distribution. Reconstruction-based approaches that use generative models to learn to reconstruct healthy brain anatomy are commonly used for this task. Diffusion models are an emerging class of deep generative models that show great potential regarding reconstruction fidelity. However, they face challenges in preserving intensity characteristics in the reconstructed images, limiting their performance in anomaly detection. To address this challenge, we propose to condition the denoising mechanism of diffusion models with additional information about the image to reconstruct coming from a latent representation of the noise-free input image. This conditioning enables high-fidelity reconstruction of healthy brain structures while aligning local intensity characteristics of input-reconstruction pairs. We evaluate our method's reconstruction quality, domain adaptation features and finally segmentation performance on publicly available data sets with various pathologies. Using our proposed conditioning mechanism we can reduce the false-positive predictions and enable a more precise delineation of anomalies which significantly enhances the anomaly detection performance compared to established state-of-the-art approaches to unsupervised anomaly detection in brain MRI. Furthermore, our approach shows promise in domain adaptation across different MRI acquisitions and simulated contrasts, a crucial property of general anomaly detection methods.
Collaborative Robotic Biopsy with Trajectory Guidance and Needle Tip Force Feedback
Jun 12, 2023Abstract:The diagnostic value of biopsies is highly dependent on the placement of needles. Robotic trajectory guidance has been shown to improve needle positioning, but feedback for real-time navigation is limited. Haptic display of needle tip forces can provide rich feedback for needle navigation by enabling localization of tissue structures along the insertion path. We present a collaborative robotic biopsy system that combines trajectory guidance with kinesthetic feedback to assist the physician in needle placement. The robot aligns the needle while the insertion is performed in collaboration with a medical expert who controls the needle position on site. We present a needle design that senses forces at the needle tip based on optical coherence tomography and machine learning for real-time data processing. Our robotic setup allows operators to sense deep tissue interfaces independent of frictional forces to improve needle placement relative to a desired target structure. We first evaluate needle tip force sensing in ex-vivo tissue in a phantom study. We characterize the tip forces during insertions with constant velocity and demonstrate the ability to detect tissue interfaces in a collaborative user study. Participants are able to detect 91% of ex-vivo tissue interfaces based on needle tip force feedback alone. Finally, we demonstrate that even smaller, deep target structures can be accurately sampled by performing post-mortem in situ biopsies of the pancreas.
Robotic Tissue Sampling for Safe Post-mortem Biopsy in Infectious Corpses
Jan 28, 2022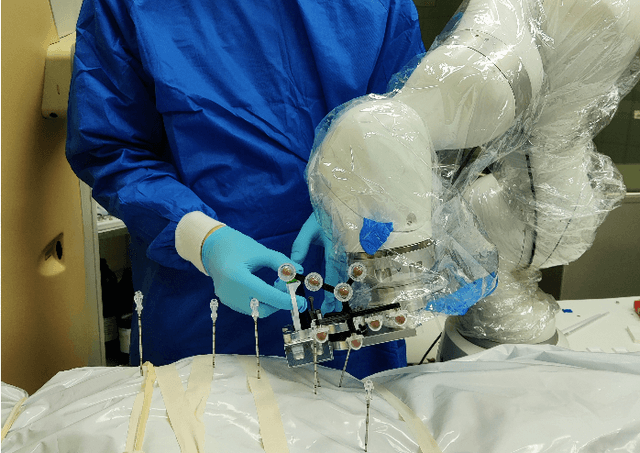
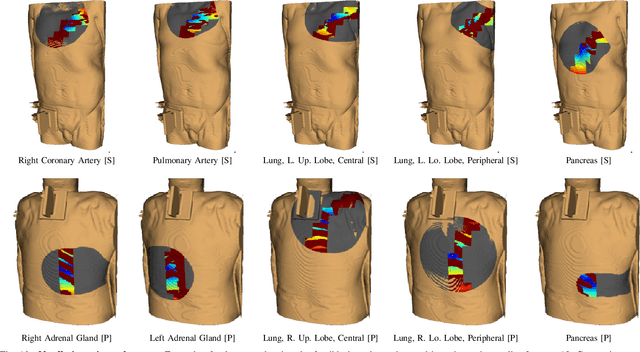
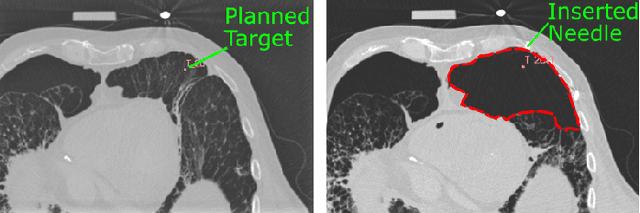
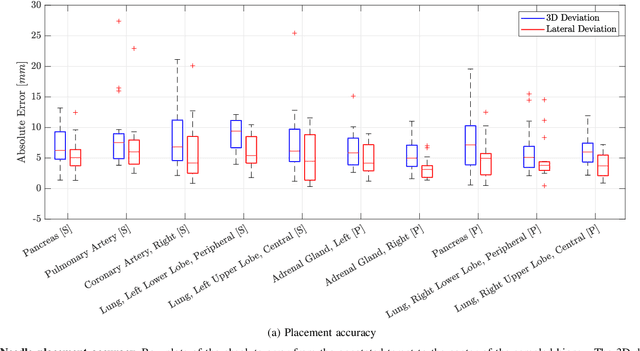
Abstract:In pathology and legal medicine, the histopathological and microbiological analysis of tissue samples from infected deceased is a valuable information for developing treatment strategies during a pandemic such as COVID-19. However, a conventional autopsy carries the risk of disease transmission and may be rejected by relatives. We propose minimally invasive biopsy with robot assistance under CT guidance to minimize the risk of disease transmission during tissue sampling and to improve accuracy. A flexible robotic system for biopsy sampling is presented, which is applied to human corpses placed inside protective body bags. An automatic planning and decision system estimates optimal insertion point. Heat maps projected onto the segmented skin visualize the distance and angle of insertions and estimate the minimum cost of a puncture while avoiding bone collisions. Further, we test multiple insertion paths concerning feasibility and collisions. A custom end effector is designed for inserting needles and extracting tissue samples under robotic guidance. Our robotic post-mortem biopsy (RPMB) system is evaluated in a study during the COVID-19 pandemic on 20 corpses and 10 tissue targets, 5 of them being infected with SARS-CoV-2. The mean planning time including robot path planning is (5.72+-1.67) s. Mean needle placement accuracy is (7.19+-4.22) mm.
A novel optical needle probe for deep learning-based tissue elasticity characterization
Sep 20, 2021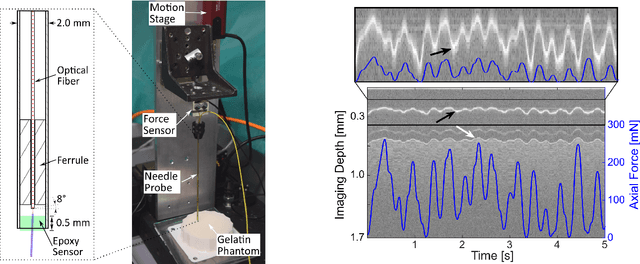
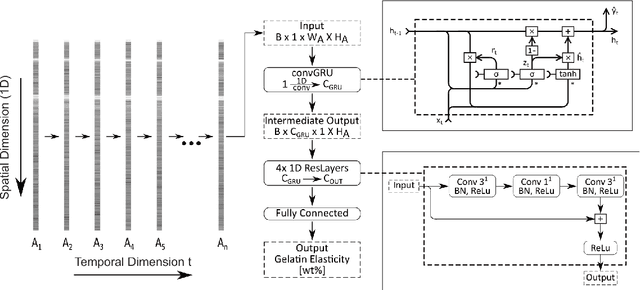
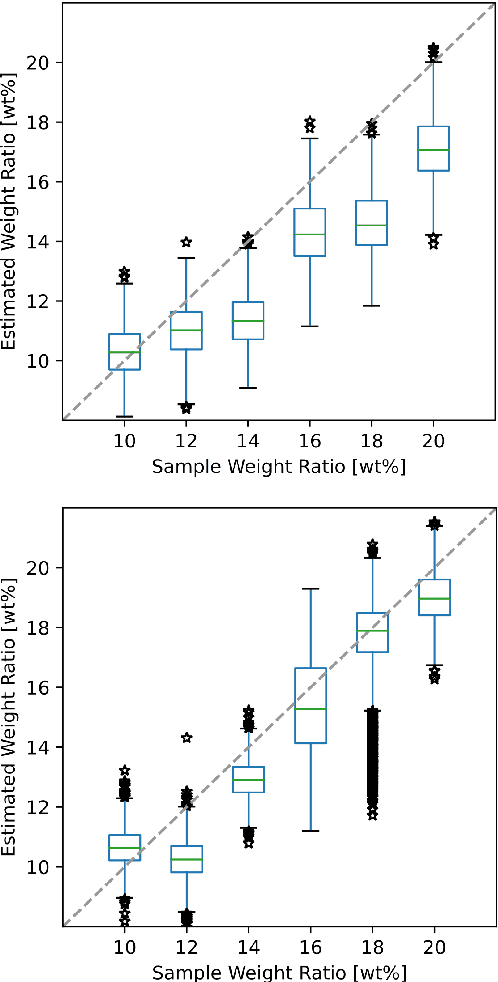
Abstract:The distinction between malignant and benign tumors is essential to the treatment of cancer. The tissue's elasticity can be used as an indicator for the required tissue characterization. Optical coherence elastography (OCE) probes have been proposed for needle insertions but have so far lacked the necessary load sensing capabilities. We present a novel OCE needle probe that provides simultaneous optical coherence tomography (OCT) imaging and load sensing at the needle tip. We demonstrate the application of the needle probe in indentation experiments on gelatin phantoms with varying gelatin concentrations. We further implement two deep learning methods for the end-to-end sample characterization from the acquired OCT data. We report the estimation of gelatin sample concentrations in unseen samples with a mean error of $1.21 \pm 0.91$ wt\%. Both evaluated deep learning models successfully provide sample characterization with different advantages regarding the accuracy and inference time.
* Accepted at CURAC 2021, 2nd Place in the Best Paper Awards
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge