Marta Ligero
Abnormality-Driven Representation Learning for Radiology Imaging
Nov 25, 2024



Abstract:To date, the most common approach for radiology deep learning pipelines is the use of end-to-end 3D networks based on models pre-trained on other tasks, followed by fine-tuning on the task at hand. In contrast, adjacent medical fields such as pathology, which focus on 2D images, have effectively adopted task-agnostic foundational models based on self-supervised learning (SSL), combined with weakly-supervised deep learning (DL). However, the field of radiology still lacks task-agnostic representation models due to the computational and data demands of 3D imaging and the anatomical complexity inherent to radiology scans. To address this gap, we propose CLEAR, a framework for radiology images that uses extracted embeddings from 2D slices along with attention-based aggregation for efficiently predicting clinical endpoints. As part of this framework, we introduce lesion-enhanced contrastive learning (LeCL), a novel approach to obtain visual representations driven by abnormalities in 2D axial slices across different locations of the CT scans. Specifically, we trained single-domain contrastive learning approaches using three different architectures: Vision Transformers, Vision State Space Models and Gated Convolutional Neural Networks. We evaluate our approach across three clinical tasks: tumor lesion location, lung disease detection, and patient staging, benchmarking against four state-of-the-art foundation models, including BiomedCLIP. Our findings demonstrate that CLEAR using representations learned through LeCL, outperforms existing foundation models, while being substantially more compute- and data-efficient.
Unsupervised Foundation Model-Agnostic Slide-Level Representation Learning
Nov 20, 2024



Abstract:Representation learning of pathology whole-slide images (WSIs) has primarily relied on weak supervision with Multiple Instance Learning (MIL). This approach leads to slide representations highly tailored to a specific clinical task. Self-supervised learning (SSL) has been successfully applied to train histopathology foundation models (FMs) for patch embedding generation. However, generating patient or slide level embeddings remains challenging. Existing approaches for slide representation learning extend the principles of SSL from patch level learning to entire slides by aligning different augmentations of the slide or by utilizing multimodal data. By integrating tile embeddings from multiple FMs, we propose a new single modality SSL method in feature space that generates useful slide representations. Our contrastive pretraining strategy, called COBRA, employs multiple FMs and an architecture based on Mamba-2. COBRA exceeds performance of state-of-the-art slide encoders on four different public CPTAC cohorts on average by at least +3.8% AUC, despite only being pretrained on 3048 WSIs from TCGA. Additionally, COBRA is readily compatible at inference time with previously unseen feature extractors.
Reducing self-supervised learning complexity improves weakly-supervised classification performance in computational pathology
Mar 12, 2024Abstract:Deep Learning models have been successfully utilized to extract clinically actionable insights from routinely available histology data. Generally, these models require annotations performed by clinicians, which are scarce and costly to generate. The emergence of self-supervised learning (SSL) methods remove this barrier, allowing for large-scale analyses on non-annotated data. However, recent SSL approaches apply increasingly expansive model architectures and larger datasets, causing the rapid escalation of data volumes, hardware prerequisites, and overall expenses, limiting access to these resources to few institutions. Therefore, we investigated the complexity of contrastive SSL in computational pathology in relation to classification performance with the utilization of consumer-grade hardware. Specifically, we analyzed the effects of adaptations in data volume, architecture, and algorithms on downstream classification tasks, emphasizing their impact on computational resources. We trained breast cancer foundation models on a large public patient cohort and validated them on various downstream classification tasks in a weakly supervised manner on two external public patient cohorts. Our experiments demonstrate that we can improve downstream classification performance whilst reducing SSL training duration by 90%. In summary, we propose a set of adaptations which enable the utilization of SSL in computational pathology in non-resource abundant environments.
In-context learning enables multimodal large language models to classify cancer pathology images
Mar 12, 2024Abstract:Medical image classification requires labeled, task-specific datasets which are used to train deep learning networks de novo, or to fine-tune foundation models. However, this process is computationally and technically demanding. In language processing, in-context learning provides an alternative, where models learn from within prompts, bypassing the need for parameter updates. Yet, in-context learning remains underexplored in medical image analysis. Here, we systematically evaluate the model Generative Pretrained Transformer 4 with Vision capabilities (GPT-4V) on cancer image processing with in-context learning on three cancer histopathology tasks of high importance: Classification of tissue subtypes in colorectal cancer, colon polyp subtyping and breast tumor detection in lymph node sections. Our results show that in-context learning is sufficient to match or even outperform specialized neural networks trained for particular tasks, while only requiring a minimal number of samples. In summary, this study demonstrates that large vision language models trained on non-domain specific data can be applied out-of-the box to solve medical image-processing tasks in histopathology. This democratizes access of generalist AI models to medical experts without technical background especially for areas where annotated data is scarce.
Joint multi-task learning improves weakly-supervised biomarker prediction in computational pathology
Mar 06, 2024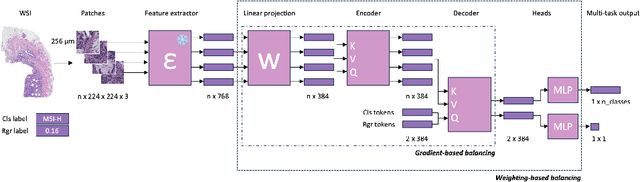


Abstract:Deep Learning (DL) can predict biomarkers directly from digitized cancer histology in a weakly-supervised setting. Recently, the prediction of continuous biomarkers through regression-based DL has seen an increasing interest. Nonetheless, clinical decision making often requires a categorical outcome. Consequently, we developed a weakly-supervised joint multi-task Transformer architecture which has been trained and evaluated on four public patient cohorts for the prediction of two key predictive biomarkers, microsatellite instability (MSI) and homologous recombination deficiency (HRD), trained with auxiliary regression tasks related to the tumor microenvironment. Moreover, we perform a comprehensive benchmark of 16 approaches of task balancing for weakly-supervised joint multi-task learning in computational pathology. Using our novel approach, we improve over the state-of-the-art area under the receiver operating characteristic by +7.7% and +4.1%, as well as yielding better clustering of latent embeddings by +8% and +5% for the prediction of MSI and HRD in external cohorts, respectively.
From Whole-slide Image to Biomarker Prediction: A Protocol for End-to-End Deep Learning in Computational Pathology
Dec 18, 2023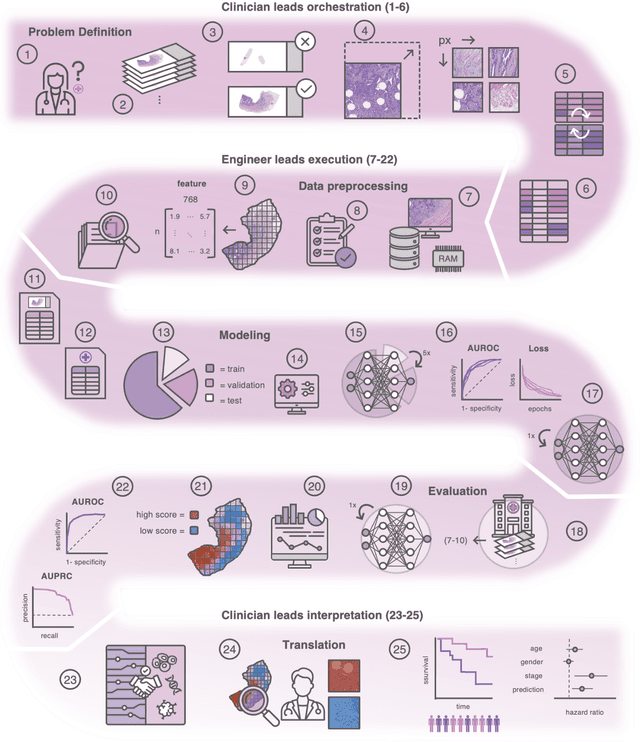
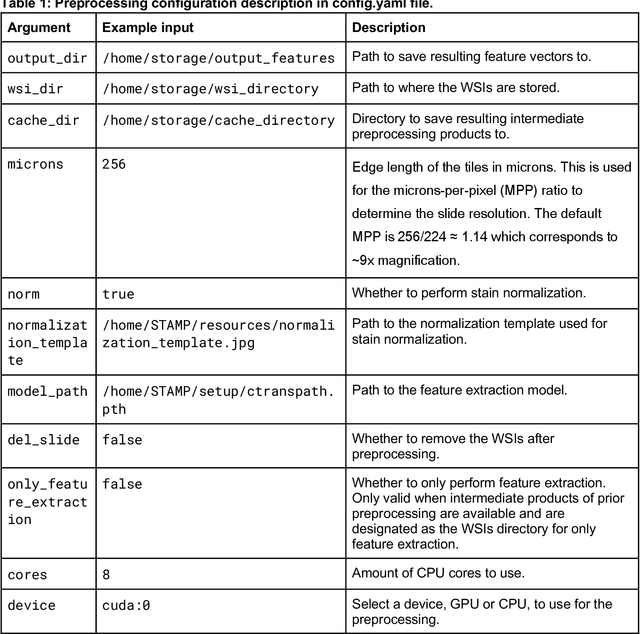
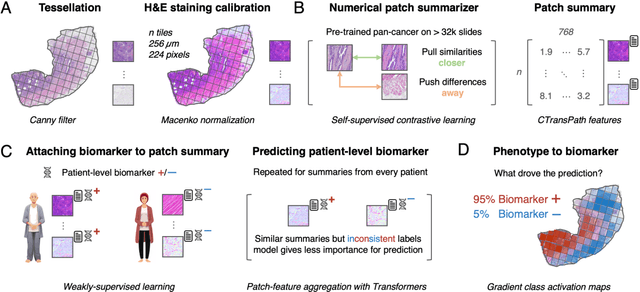

Abstract:Hematoxylin- and eosin (H&E) stained whole-slide images (WSIs) are the foundation of diagnosis of cancer. In recent years, development of deep learning-based methods in computational pathology enabled the prediction of biomarkers directly from WSIs. However, accurately linking tissue phenotype to biomarkers at scale remains a crucial challenge for democratizing complex biomarkers in precision oncology. This protocol describes a practical workflow for solid tumor associative modeling in pathology (STAMP), enabling prediction of biomarkers directly from WSIs using deep learning. The STAMP workflow is biomarker agnostic and allows for genetic- and clinicopathologic tabular data to be included as an additional input, together with histopathology images. The protocol consists of five main stages which have been successfully applied to various research problems: formal problem definition, data preprocessing, modeling, evaluation and clinical translation. The STAMP workflow differentiates itself through its focus on serving as a collaborative framework that can be used by clinicians and engineers alike for setting up research projects in the field of computational pathology. As an example task, we applied STAMP to the prediction of microsatellite instability (MSI) status in colorectal cancer, showing accurate performance for the identification of MSI-high tumors. Moreover, we provide an open-source codebase which has been deployed at several hospitals across the globe to set up computational pathology workflows. The STAMP workflow requires one workday of hands-on computational execution and basic command line knowledge.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge