David J. Harrison
SurGen: 1020 H&E-stained Whole Slide Images With Survival and Genetic Markers
Feb 07, 2025Abstract:$\textbf{Background}$: Cancer remains one of the leading causes of morbidity and mortality worldwide. Comprehensive datasets that combine histopathological images with genetic and survival data across various tumour sites are essential for advancing computational pathology and personalised medicine. $\textbf{Results}$: We present SurGen, a dataset comprising 1,020 H&E-stained whole slide images (WSIs) from 843 colorectal cancer cases. The dataset includes detailed annotations for key genetic mutations (KRAS, NRAS, BRAF) and mismatch repair status, as well as survival data for 426 cases. To demonstrate SurGen's practical utility, we conducted a proof-of-concept machine learning experiment predicting mismatch repair status from the WSIs, achieving a test AUROC of 0.8316. These preliminary results underscore the dataset's potential to facilitate research in biomarker discovery, prognostic modelling, and advanced machine learning applications in colorectal cancer. $\textbf{Conclusions}$: SurGen offers a valuable resource for the scientific community, enabling studies that require high-quality WSIs linked with comprehensive clinical and genetic information on colorectal cancer. Our initial findings affirm the dataset's capacity to advance diagnostic precision and foster the development of personalised treatment strategies in colorectal oncology. Data available online at https://doi.org/10.6019/S-BIAD1285.
A Good Feature Extractor Is All You Need for Weakly Supervised Learning in Histopathology
Nov 29, 2023Abstract:Deep learning is revolutionising pathology, offering novel opportunities in disease prognosis and personalised treatment. Historically, stain normalisation has been a crucial preprocessing step in computational pathology pipelines, and persists into the deep learning era. Yet, with the emergence of feature extractors trained using self-supervised learning (SSL) on diverse pathology datasets, we call this practice into question. In an empirical evaluation of publicly available feature extractors, we find that omitting stain normalisation and image augmentations does not compromise downstream performance, while incurring substantial savings in memory and compute. Further, we show that the top-performing feature extractors are remarkably robust to variations in stain and augmentations like rotation in their latent space. Contrary to previous patch-level benchmarking studies, our approach emphasises clinical relevance by focusing on slide-level prediction tasks in a weakly supervised setting with external validation cohorts. This work represents the most comprehensive robustness evaluation of public pathology SSL feature extractors to date, involving more than 6,000 training runs across nine tasks, five datasets, three downstream architectures, and various preprocessing setups. Our findings stand to streamline digital pathology workflows by minimising preprocessing needs and informing the selection of feature extractors.
Deep Multiple Instance Learning with Distance-Aware Self-Attention
May 20, 2023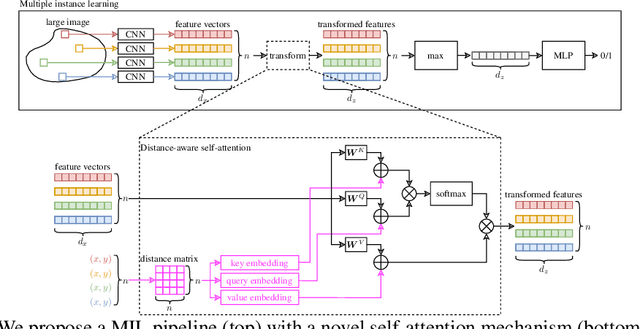
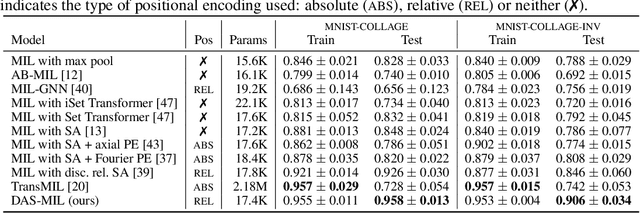
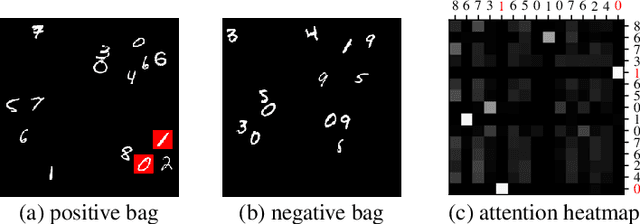
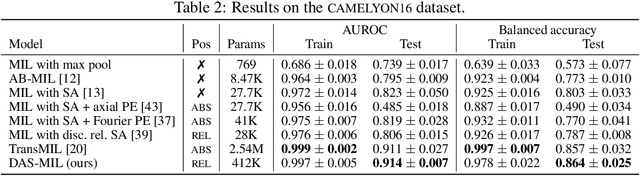
Abstract:Traditional supervised learning tasks require a label for every instance in the training set, but in many real-world applications, labels are only available for collections (bags) of instances. This problem setting, known as multiple instance learning (MIL), is particularly relevant in the medical domain, where high-resolution images are split into smaller patches, but labels apply to the image as a whole. Recent MIL models are able to capture correspondences between patches by employing self-attention, allowing them to weigh each patch differently based on all other patches in the bag. However, these approaches still do not consider the relative spatial relationships between patches within the larger image, which is especially important in computational pathology. To this end, we introduce a novel MIL model with distance-aware self-attention (DAS-MIL), which explicitly takes into account relative spatial information when modelling the interactions between patches. Unlike existing relative position representations for self-attention which are discrete, our approach introduces continuous distance-dependent terms into the computation of the attention weights, and is the first to apply relative position representations in the context of MIL. We evaluate our model on a custom MNIST-based MIL dataset that requires the consideration of relative spatial information, as well as on CAMELYON16, a publicly available cancer metastasis detection dataset, where we achieve a test AUROC score of 0.91. On both datasets, our model outperforms existing MIL approaches that employ absolute positional encodings, as well as existing relative position representation schemes applied to MIL. Our code is available at https://anonymous.4open.science/r/das-mil.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge