Danush Kumar Venkatesh
Mission Balance: Generating Under-represented Class Samples using Video Diffusion Models
May 14, 2025Abstract:Computer-assisted interventions can improve intra-operative guidance, particularly through deep learning methods that harness the spatiotemporal information in surgical videos. However, the severe data imbalance often found in surgical video datasets hinders the development of high-performing models. In this work, we aim to overcome the data imbalance by synthesizing surgical videos. We propose a unique two-stage, text-conditioned diffusion-based method to generate high-fidelity surgical videos for under-represented classes. Our approach conditions the generation process on text prompts and decouples spatial and temporal modeling by utilizing a 2D latent diffusion model to capture spatial content and then integrating temporal attention layers to ensure temporal consistency. Furthermore, we introduce a rejection sampling strategy to select the most suitable synthetic samples, effectively augmenting existing datasets to address class imbalance. We evaluate our method on two downstream tasks-surgical action recognition and intra-operative event prediction-demonstrating that incorporating synthetic videos from our approach substantially enhances model performance. We open-source our implementation at https://gitlab.com/nct_tso_public/surgvgen.
Synthesizing Multi-Class Surgical Datasets with Anatomy-Aware Diffusion Models
Oct 10, 2024Abstract:In computer-assisted surgery, automatically recognizing anatomical organs is crucial for understanding the surgical scene and providing intraoperative assistance. While machine learning models can identify such structures, their deployment is hindered by the need for labeled, diverse surgical datasets with anatomical annotations. Labeling multiple classes (i.e., organs) in a surgical scene is time-intensive, requiring medical experts. Although synthetically generated images can enhance segmentation performance, maintaining both organ structure and texture during generation is challenging. We introduce a multi-stage approach using diffusion models to generate multi-class surgical datasets with annotations. Our framework improves anatomy awareness by training organ specific models with an inpainting objective guided by binary segmentation masks. The organs are generated with an inference pipeline using pre-trained ControlNet to maintain the organ structure. The synthetic multi-class datasets are constructed through an image composition step, ensuring structural and textural consistency. This versatile approach allows the generation of multi-class datasets from real binary datasets and simulated surgical masks. We thoroughly evaluate the generated datasets on image quality and downstream segmentation, achieving a $15\%$ improvement in segmentation scores when combined with real images. Our codebase https://gitlab.com/nct_tso_public/muli-class-image-synthesis
SurgicaL-CD: Generating Surgical Images via Unpaired Image Translation with Latent Consistency Diffusion Models
Aug 19, 2024Abstract:Computer-assisted surgery (CAS) systems are designed to assist surgeons during procedures, thereby reducing complications and enhancing patient care. Training machine learning models for these systems requires a large corpus of annotated datasets, which is challenging to obtain in the surgical domain due to patient privacy concerns and the significant labeling effort required from doctors. Previous methods have explored unpaired image translation using generative models to create realistic surgical images from simulations. However, these approaches have struggled to produce high-quality, diverse surgical images. In this work, we introduce \emph{SurgicaL-CD}, a consistency-distilled diffusion method to generate realistic surgical images with only a few sampling steps without paired data. We evaluate our approach on three datasets, assessing the generated images in terms of quality and utility as downstream training datasets. Our results demonstrate that our method outperforms GANs and diffusion-based approaches. Our code is available at \url{https://gitlab.com/nct_tso_public/gan2diffusion}.
Exploring Semantic Consistency in Unpaired Image Translation to Generate Data for Surgical Applications
Sep 11, 2023Abstract:In surgical computer vision applications, obtaining labeled training data is challenging due to data-privacy concerns and the need for expert annotation. Unpaired image-to-image translation techniques have been explored to automatically generate large annotated datasets by translating synthetic images to the realistic domain. However, preserving the structure and semantic consistency between the input and translated images presents significant challenges, mainly when there is a distributional mismatch in the semantic characteristics of the domains. This study empirically investigates unpaired image translation methods for generating suitable data in surgical applications, explicitly focusing on semantic consistency. We extensively evaluate various state-of-the-art image translation models on two challenging surgical datasets and downstream semantic segmentation tasks. We find that a simple combination of structural-similarity loss and contrastive learning yields the most promising results. Quantitatively, we show that the data generated with this approach yields higher semantic consistency and can be used more effectively as training data.
Detecting Adversarial Examples in Batches -- a geometrical approach
Jun 17, 2022

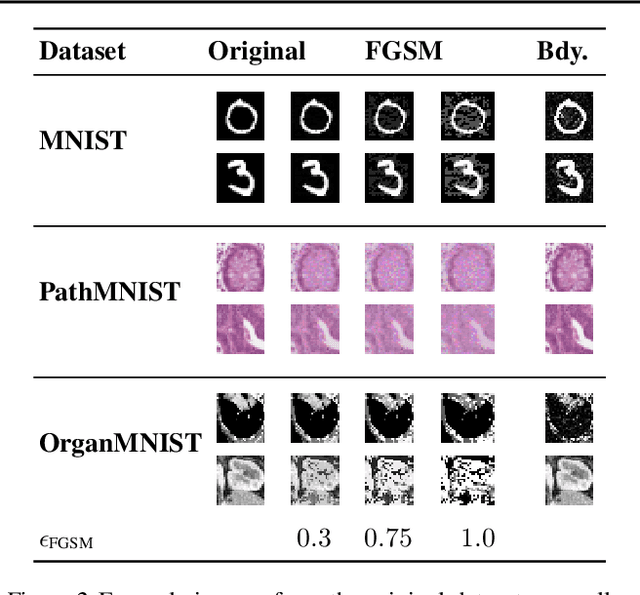

Abstract:Many deep learning methods have successfully solved complex tasks in computer vision and speech recognition applications. Nonetheless, the robustness of these models has been found to be vulnerable to perturbed inputs or adversarial examples, which are imperceptible to the human eye, but lead the model to erroneous output decisions. In this study, we adapt and introduce two geometric metrics, density and coverage, and evaluate their use in detecting adversarial samples in batches of unseen data. We empirically study these metrics using MNIST and two real-world biomedical datasets from MedMNIST, subjected to two different adversarial attacks. Our experiments show promising results for both metrics to detect adversarial examples. We believe that his work can lay the ground for further study on these metrics' use in deployed machine learning systems to monitor for possible attacks by adversarial examples or related pathologies such as dataset shift.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge