Steffen Löck
Unraveling Radiomics Complexity: Strategies for Optimal Simplicity in Predictive Modeling
Jul 05, 2024



Abstract:Background: The high dimensionality of radiomic feature sets, the variability in radiomic feature types and potentially high computational requirements all underscore the need for an effective method to identify the smallest set of predictive features for a given clinical problem. Purpose: Develop a methodology and tools to identify and explain the smallest set of predictive radiomic features. Materials and Methods: 89,714 radiomic features were extracted from five cancer datasets: low-grade glioma, meningioma, non-small cell lung cancer (NSCLC), and two renal cell carcinoma cohorts (n=2104). Features were categorized by computational complexity into morphological, intensity, texture, linear filters, and nonlinear filters. Models were trained and evaluated on each complexity level using the area under the curve (AUC). The most informative features were identified, and their importance was explained. The optimal complexity level and associated most informative features were identified using systematic statistical significance analyses and a false discovery avoidance procedure, respectively. Their predictive importance was explained using a novel tree-based method. Results: MEDimage, a new open-source tool, was developed to facilitate radiomic studies. Morphological features were optimal for MRI-based meningioma (AUC: 0.65) and low-grade glioma (AUC: 0.68). Intensity features were optimal for CECT-based renal cell carcinoma (AUC: 0.82) and CT-based NSCLC (AUC: 0.76). Texture features were optimal for MRI-based renal cell carcinoma (AUC: 0.72). Tuning the Hounsfield unit range improved results for CECT-based renal cell carcinoma (AUC: 0.86). Conclusion: Our proposed methodology and software can estimate the optimal radiomics complexity level for specific medical outcomes, potentially simplifying the use of radiomics in predictive modeling across various contexts.
Graph data modelling for outcome prediction in oropharyngeal cancer patients
Oct 04, 2023



Abstract:Graph neural networks (GNNs) are becoming increasingly popular in the medical domain for the tasks of disease classification and outcome prediction. Since patient data is not readily available as a graph, most existing methods either manually define a patient graph, or learn a latent graph based on pairwise similarities between the patients. There are also hypergraph neural network (HGNN)-based methods that were introduced recently to exploit potential higher order associations between the patients by representing them as a hypergraph. In this work, we propose a patient hypergraph network (PHGN), which has been investigated in an inductive learning setup for binary outcome prediction in oropharyngeal cancer (OPC) patients using computed tomography (CT)-based radiomic features for the first time. Additionally, the proposed model was extended to perform time-to-event analyses, and compared with GNN and baseline linear models.
Image biomarker standardisation initiative
Sep 17, 2018
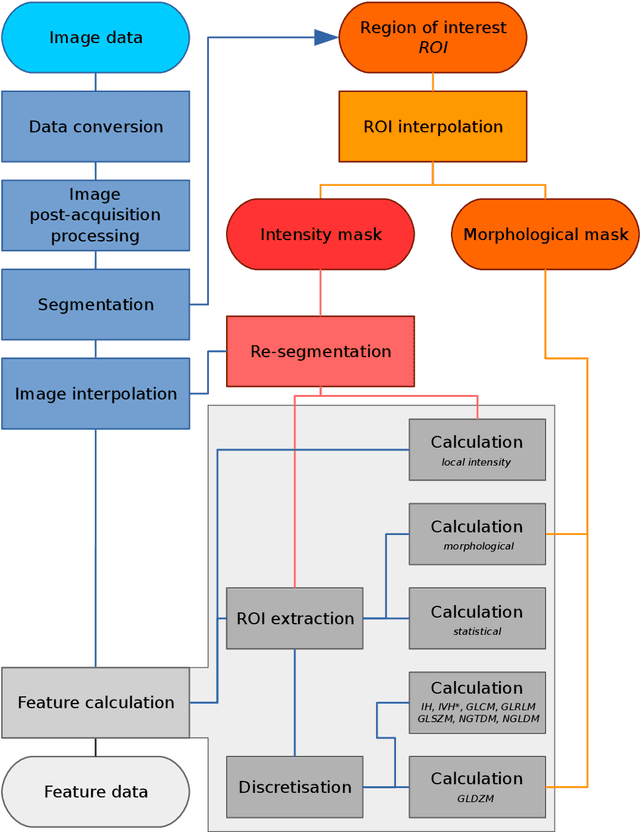
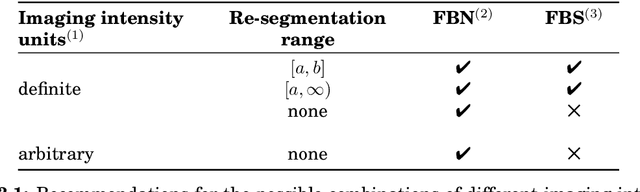
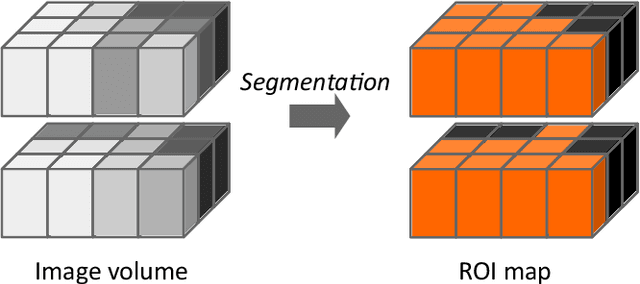
Abstract:The image biomarker standardisation initiative (IBSI) is an independent international collaboration which works towards standardising the extraction of image biomarkers from acquired imaging for the purpose of high-throughput quantitative image analysis (radiomics). Lack of reproducibility and validation of high-throughput quantitative image analysis studies is considered to be a major challenge for the field. Part of this challenge lies in the scantiness of consensus-based guidelines and definitions for the process of translating acquired imaging into high-throughput image biomarkers. The IBSI therefore seeks to provide image biomarker nomenclature and definitions, benchmark data sets, and benchmark values to verify image processing and image biomarker calculations, as well as reporting guidelines, for high-throughput image analysis.
Assessing robustness of radiomic features by image perturbation
Jun 18, 2018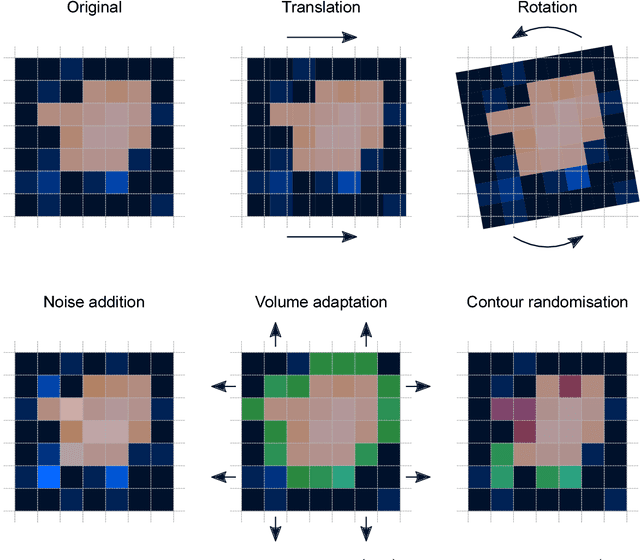
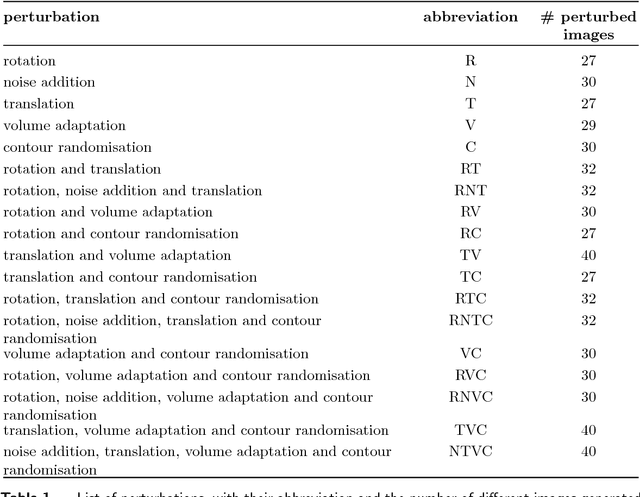
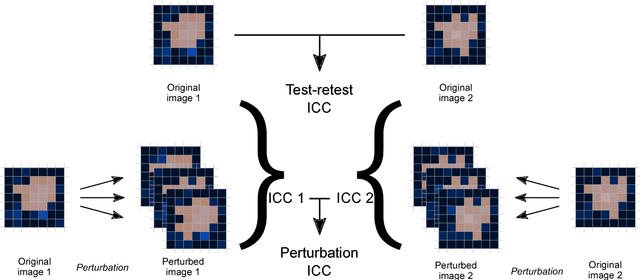
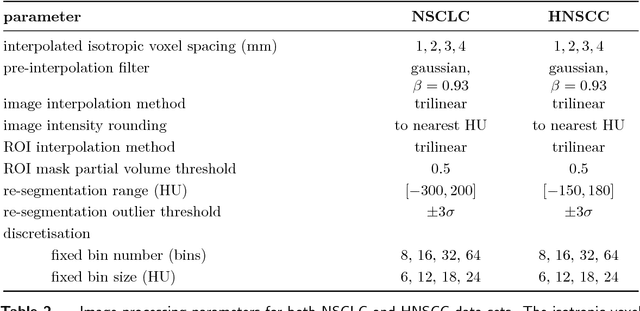
Abstract:Image features need to be robust against differences in positioning, acquisition and segmentation to ensure reproducibility. Radiomic models that only include robust features can be used to analyse new images, whereas models with non-robust features may fail to predict the outcome of interest accurately. Test-retest imaging is recommended to assess robustness, but may not be available for the phenotype of interest. We therefore investigated 18 methods to determine feature robustness based on image perturbations. Test-retest and perturbation robustness were compared for 4032 features that were computed from the gross tumour volume in two cohorts with computed tomography imaging: I) 31 non-small-cell lung cancer (NSCLC) patients; II): 19 head-and-neck squamous cell carcinoma (HNSCC) patients. Robustness was measured using the intraclass correlation coefficient (1,1) (ICC). Features with ICC$\geq0.90$ were considered robust. The NSCLC cohort contained more robust features for test-retest imaging than the HNSCC cohort ($73.5\%$ vs. $34.0\%$). A perturbation chain consisting of noise addition, affine translation, volume growth/shrinkage and supervoxel-based contour randomisation identified the fewest false positive robust features (NSCLC: $3.3\%$; HNSCC: $10.0\%$). Thus, this perturbation chain may be used to assess feature robustness.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge