Xukun Zhang
VLM-based Prompts as the Optimal Assistant for Unpaired Histopathology Virtual Staining
Apr 22, 2025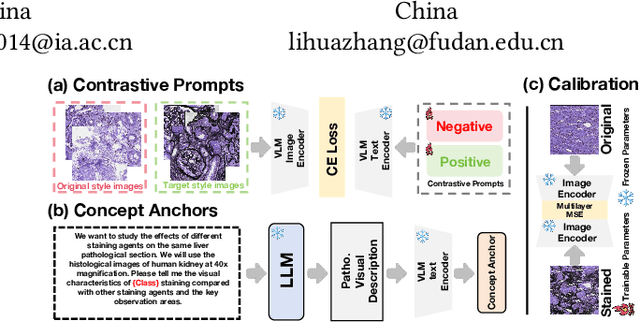
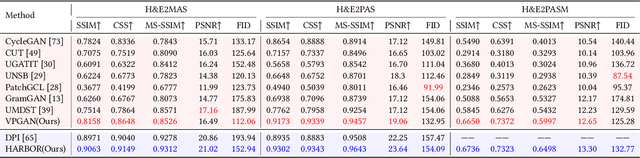
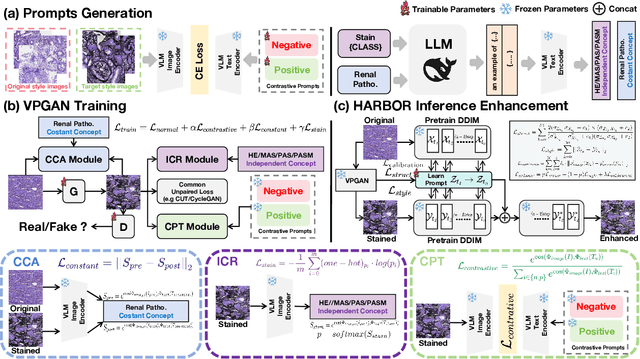
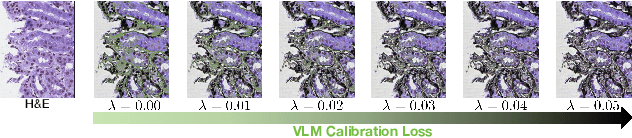
Abstract:In histopathology, tissue sections are typically stained using common H&E staining or special stains (MAS, PAS, PASM, etc.) to clearly visualize specific tissue structures. The rapid advancement of deep learning offers an effective solution for generating virtually stained images, significantly reducing the time and labor costs associated with traditional histochemical staining. However, a new challenge arises in separating the fundamental visual characteristics of tissue sections from the visual differences induced by staining agents. Additionally, virtual staining often overlooks essential pathological knowledge and the physical properties of staining, resulting in only style-level transfer. To address these issues, we introduce, for the first time in virtual staining tasks, a pathological vision-language large model (VLM) as an auxiliary tool. We integrate contrastive learnable prompts, foundational concept anchors for tissue sections, and staining-specific concept anchors to leverage the extensive knowledge of the pathological VLM. This approach is designed to describe, frame, and enhance the direction of virtual staining. Furthermore, we have developed a data augmentation method based on the constraints of the VLM. This method utilizes the VLM's powerful image interpretation capabilities to further integrate image style and structural information, proving beneficial in high-precision pathological diagnostics. Extensive evaluations on publicly available multi-domain unpaired staining datasets demonstrate that our method can generate highly realistic images and enhance the accuracy of downstream tasks, such as glomerular detection and segmentation. Our code is available at: https://github.com/CZZZZZZZZZZZZZZZZZ/VPGAN-HARBOR
VGAT: A Cancer Survival Analysis Framework Transitioning from Generative Visual Question Answering to Genomic Reconstruction
Mar 25, 2025Abstract:Multimodal learning combining pathology images and genomic sequences enhances cancer survival analysis but faces clinical implementation barriers due to limited access to genomic sequencing in under-resourced regions. To enable survival prediction using only whole-slide images (WSI), we propose the Visual-Genomic Answering-Guided Transformer (VGAT), a framework integrating Visual Question Answering (VQA) techniques for genomic modality reconstruction. By adapting VQA's text feature extraction approach, we derive stable genomic representations that circumvent dimensionality challenges in raw genomic data. Simultaneously, a cluster-based visual prompt module selectively enhances discriminative WSI patches, addressing noise from unfiltered image regions. Evaluated across five TCGA datasets, VGAT outperforms existing WSI-only methods, demonstrating the viability of genomic-informed inference without sequencing. This approach bridges multimodal research and clinical feasibility in resource-constrained settings. The code link is https://github.com/CZZZZZZZZZZZZZZZZZ/VGAT.
Towards Unified Molecule-Enhanced Pathology Image Representation Learning via Integrating Spatial Transcriptomics
Dec 01, 2024



Abstract:Recent advancements in multimodal pre-training models have significantly advanced computational pathology. However, current approaches predominantly rely on visual-language models, which may impose limitations from a molecular perspective and lead to performance bottlenecks. Here, we introduce a Unified Molecule-enhanced Pathology Image REpresentationn Learning framework (UMPIRE). UMPIRE aims to leverage complementary information from gene expression profiles to guide the multimodal pre-training, enhancing the molecular awareness of pathology image representation learning. We demonstrate that this molecular perspective provides a robust, task-agnostic training signal for learning pathology image embeddings. Due to the scarcity of paired data, approximately 4 million entries of spatial transcriptomics gene expression were collected to train the gene encoder. By leveraging powerful pre-trained encoders, UMPIRE aligns the encoders across over 697K pathology image-gene expression pairs. The performance of UMPIRE is demonstrated across various molecular-related downstream tasks, including gene expression prediction, spot classification, and mutation state prediction in whole slide images. Our findings highlight the effectiveness of multimodal data integration and open new avenues for exploring computational pathology enhanced by molecular perspectives. The code and pre-trained weights are available at https://github.com/Hanminghao/UMPIRE.
MSCPT: Few-shot Whole Slide Image Classification with Multi-scale and Context-focused Prompt Tuning
Aug 21, 2024



Abstract:Multiple instance learning (MIL) has become a standard paradigm for weakly supervised classification of whole slide images (WSI). However, this paradigm relies on the use of a large number of labelled WSIs for training. The lack of training data and the presence of rare diseases present significant challenges for these methods. Prompt tuning combined with the pre-trained Vision-Language models (VLMs) is an effective solution to the Few-shot Weakly Supervised WSI classification (FSWC) tasks. Nevertheless, applying prompt tuning methods designed for natural images to WSIs presents three significant challenges: 1) These methods fail to fully leverage the prior knowledge from the VLM's text modality; 2) They overlook the essential multi-scale and contextual information in WSIs, leading to suboptimal results; and 3) They lack exploration of instance aggregation methods. To address these problems, we propose a Multi-Scale and Context-focused Prompt Tuning (MSCPT) method for FSWC tasks. Specifically, MSCPT employs the frozen large language model to generate pathological visual language prior knowledge at multi-scale, guiding hierarchical prompt tuning. Additionally, we design a graph prompt tuning module to learn essential contextual information within WSI, and finally, a non-parametric cross-guided instance aggregation module has been introduced to get the WSI-level features. Based on two VLMs, extensive experiments and visualizations on three datasets demonstrated the powerful performance of our MSCPT.
HybridOcc: NeRF Enhanced Transformer-based Multi-Camera 3D Occupancy Prediction
Aug 17, 2024



Abstract:Vision-based 3D semantic scene completion (SSC) describes autonomous driving scenes through 3D volume representations. However, the occlusion of invisible voxels by scene surfaces poses challenges to current SSC methods in hallucinating refined 3D geometry. This paper proposes HybridOcc, a hybrid 3D volume query proposal method generated by Transformer framework and NeRF representation and refined in a coarse-to-fine SSC prediction framework. HybridOcc aggregates contextual features through the Transformer paradigm based on hybrid query proposals while combining it with NeRF representation to obtain depth supervision. The Transformer branch contains multiple scales and uses spatial cross-attention for 2D to 3D transformation. The newly designed NeRF branch implicitly infers scene occupancy through volume rendering, including visible and invisible voxels, and explicitly captures scene depth rather than generating RGB color. Furthermore, we present an innovative occupancy-aware ray sampling method to orient the SSC task instead of focusing on the scene surface, further improving the overall performance. Extensive experiments on nuScenes and SemanticKITTI datasets demonstrate the effectiveness of our HybridOcc on the SSC task.
MaskBEV: Towards A Unified Framework for BEV Detection and Map Segmentation
Aug 17, 2024
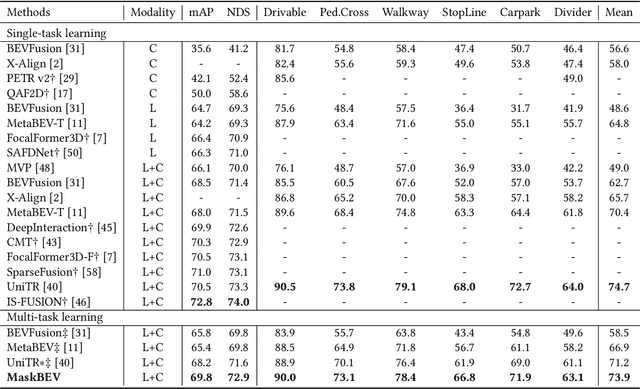

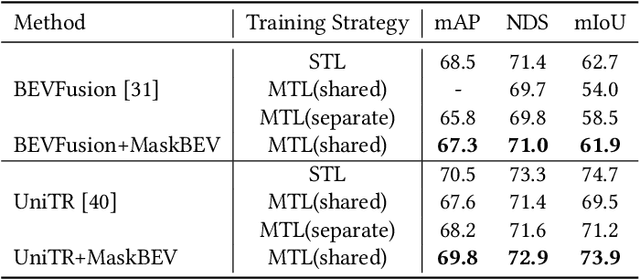
Abstract:Accurate and robust multimodal multi-task perception is crucial for modern autonomous driving systems. However, current multimodal perception research follows independent paradigms designed for specific perception tasks, leading to a lack of complementary learning among tasks and decreased performance in multi-task learning (MTL) due to joint training. In this paper, we propose MaskBEV, a masked attention-based MTL paradigm that unifies 3D object detection and bird's eye view (BEV) map segmentation. MaskBEV introduces a task-agnostic Transformer decoder to process these diverse tasks, enabling MTL to be completed in a unified decoder without requiring additional design of specific task heads. To fully exploit the complementary information between BEV map segmentation and 3D object detection tasks in BEV space, we propose spatial modulation and scene-level context aggregation strategies. These strategies consider the inherent dependencies between BEV segmentation and 3D detection, naturally boosting MTL performance. Extensive experiments on nuScenes dataset show that compared with previous state-of-the-art MTL methods, MaskBEV achieves 1.3 NDS improvement in 3D object detection and 2.7 mIoU improvement in BEV map segmentation, while also demonstrating slightly leading inference speed.
Multi-Scale Heterogeneity-Aware Hypergraph Representation for Histopathology Whole Slide Images
Apr 30, 2024Abstract:Survival prediction is a complex ordinal regression task that aims to predict the survival coefficient ranking among a cohort of patients, typically achieved by analyzing patients' whole slide images. Existing deep learning approaches mainly adopt multiple instance learning or graph neural networks under weak supervision. Most of them are unable to uncover the diverse interactions between different types of biological entities(\textit{e.g.}, cell cluster and tissue block) across multiple scales, while such interactions are crucial for patient survival prediction. In light of this, we propose a novel multi-scale heterogeneity-aware hypergraph representation framework. Specifically, our framework first constructs a multi-scale heterogeneity-aware hypergraph and assigns each node with its biological entity type. It then mines diverse interactions between nodes on the graph structure to obtain a global representation. Experimental results demonstrate that our method outperforms state-of-the-art approaches on three benchmark datasets. Code is publicly available at \href{https://github.com/Hanminghao/H2GT}{https://github.com/Hanminghao/H2GT}.
An objective comparison of methods for augmented reality in laparoscopic liver resection by preoperative-to-intraoperative image fusion
Feb 07, 2024



Abstract:Augmented reality for laparoscopic liver resection is a visualisation mode that allows a surgeon to localise tumours and vessels embedded within the liver by projecting them on top of a laparoscopic image. Preoperative 3D models extracted from CT or MRI data are registered to the intraoperative laparoscopic images during this process. In terms of 3D-2D fusion, most of the algorithms make use of anatomical landmarks to guide registration. These landmarks include the liver's inferior ridge, the falciform ligament, and the occluding contours. They are usually marked by hand in both the laparoscopic image and the 3D model, which is time-consuming and may contain errors if done by a non-experienced user. Therefore, there is a need to automate this process so that augmented reality can be used effectively in the operating room. We present the Preoperative-to-Intraoperative Laparoscopic Fusion Challenge (P2ILF), held during the Medical Imaging and Computer Assisted Interventions (MICCAI 2022) conference, which investigates the possibilities of detecting these landmarks automatically and using them in registration. The challenge was divided into two tasks: 1) A 2D and 3D landmark detection task and 2) a 3D-2D registration task. The teams were provided with training data consisting of 167 laparoscopic images and 9 preoperative 3D models from 9 patients, with the corresponding 2D and 3D landmark annotations. A total of 6 teams from 4 countries participated, whose proposed methods were evaluated on 16 images and two preoperative 3D models from two patients. All the teams proposed deep learning-based methods for the 2D and 3D landmark segmentation tasks and differentiable rendering-based methods for the registration task. Based on the experimental outcomes, we propose three key hypotheses that determine current limitations and future directions for research in this domain.
Tissue Segmentation of Thick-Slice Fetal Brain MR Scans with Guidance from High-Quality Isotropic Volumes
Aug 13, 2023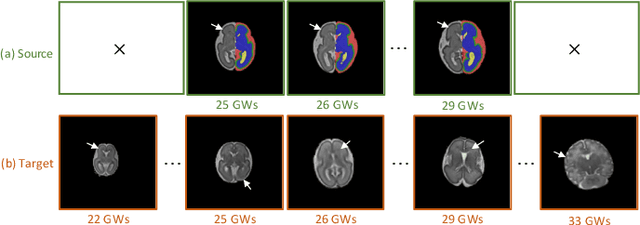
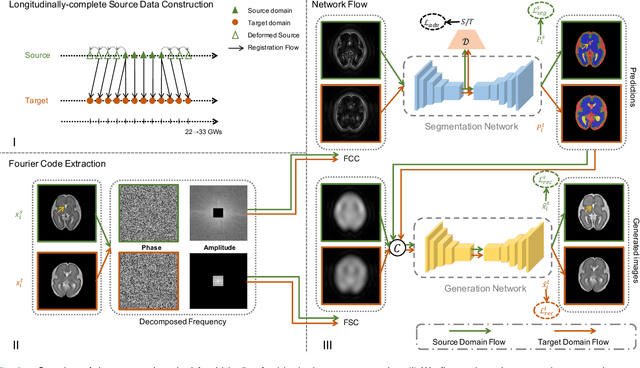
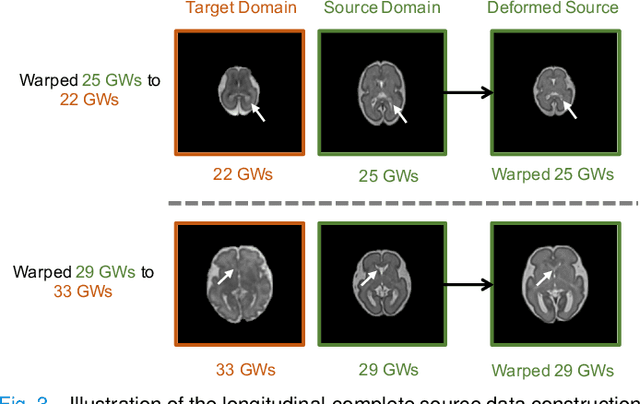
Abstract:Accurate tissue segmentation of thick-slice fetal brain magnetic resonance (MR) scans is crucial for both reconstruction of isotropic brain MR volumes and the quantification of fetal brain development. However, this task is challenging due to the use of thick-slice scans in clinically-acquired fetal brain data. To address this issue, we propose to leverage high-quality isotropic fetal brain MR volumes (and also their corresponding annotations) as guidance for segmentation of thick-slice scans. Due to existence of significant domain gap between high-quality isotropic volume (i.e., source data) and thick-slice scans (i.e., target data), we employ a domain adaptation technique to achieve the associated knowledge transfer (from high-quality <source> volumes to thick-slice <target> scans). Specifically, we first register the available high-quality isotropic fetal brain MR volumes across different gestational weeks to construct longitudinally-complete source data. To capture domain-invariant information, we then perform Fourier decomposition to extract image content and style codes. Finally, we propose a novel Cycle-Consistent Domain Adaptation Network (C2DA-Net) to efficiently transfer the knowledge learned from high-quality isotropic volumes for accurate tissue segmentation of thick-slice scans. Our C2DA-Net can fully utilize a small set of annotated isotropic volumes to guide tissue segmentation on unannotated thick-slice scans. Extensive experiments on a large-scale dataset of 372 clinically acquired thick-slice MR scans demonstrate that our C2DA-Net achieves much better performance than cutting-edge methods quantitatively and qualitatively.
Pyramid Focusing Network for mutation prediction and classification in CT images
Apr 13, 2020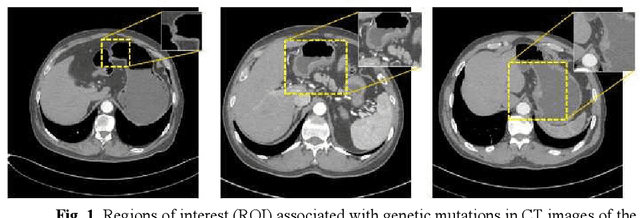
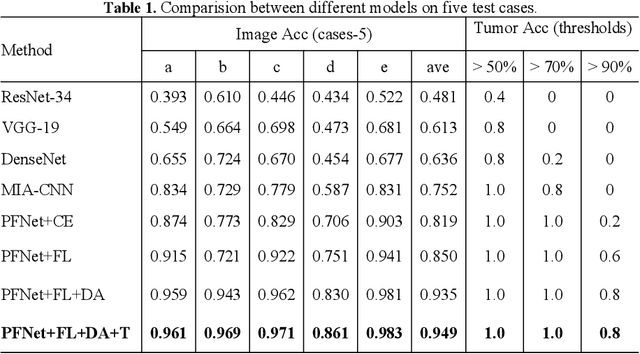
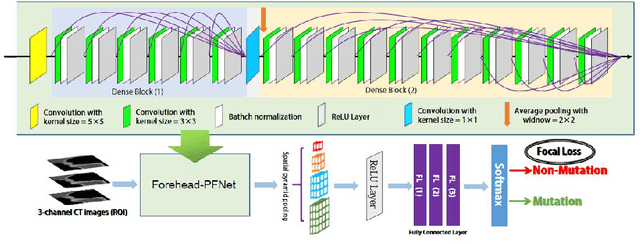
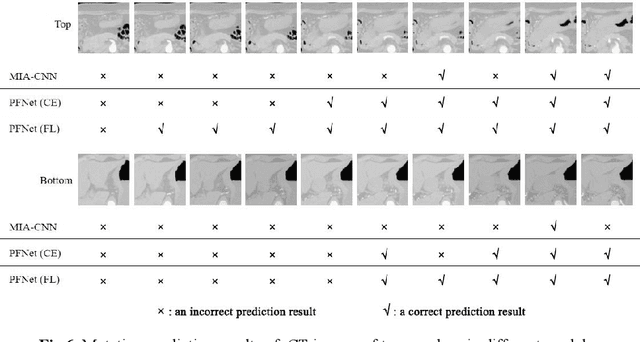
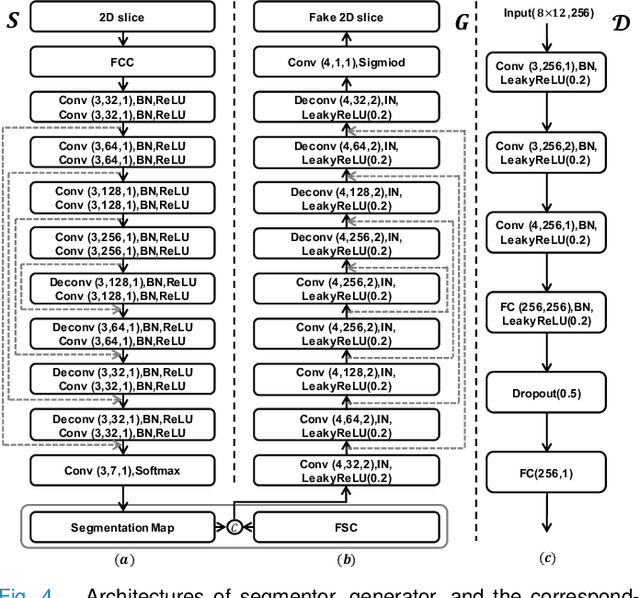
Abstract:Predicting the mutation status of genes in tumors is of great clinical significance. Recent studies have suggested that certain mutations may be noninvasively predicted by studying image features of the tumors from Computed Tomography (CT) data. Currently, this kind of image feature identification method mainly relies on manual processing to extract generalized image features alone or machine processing without considering the morphological differences of the tumor itself, which makes it difficult to achieve further breakthroughs. In this paper, we propose a pyramid focusing network (PFNet) for mutation prediction and classification based on CT images. Firstly, we use Space Pyramid Pooling to collect semantic cues in feature maps from multiple scales according to the observation that the shape and size of the tumors are varied.Secondly, we improve the loss function based on the consideration that the features required for proper mutation detection are often not obvious in cross-sections of tumor edges, which raises more attention to these hard examples in the network. Finally, we devise a training scheme based on data augmentation to enhance the generalization ability of networks. Extensively verified on clinical gastric CT datasets of 20 testing volumes with 63648 CT images, our method achieves the accuracy of 94.90% in predicting the HER-2 genes mutation status of at the CT image.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge