Linhao Qu
HUR-MACL: High-Uncertainty Region-Guided Multi-Architecture Collaborative Learning for Head and Neck Multi-Organ Segmentation
Jan 08, 2026Abstract:Accurate segmentation of organs at risk in the head and neck is essential for radiation therapy, yet deep learning models often fail on small, complexly shaped organs. While hybrid architectures that combine different models show promise, they typically just concatenate features without exploiting the unique strengths of each component. This results in functional overlap and limited segmentation accuracy. To address these issues, we propose a high uncertainty region-guided multi-architecture collaborative learning (HUR-MACL) model for multi-organ segmentation in the head and neck. This model adaptively identifies high uncertainty regions using a convolutional neural network, and for these regions, Vision Mamba as well as Deformable CNN are utilized to jointly improve their segmentation accuracy. Additionally, a heterogeneous feature distillation loss was proposed to promote collaborative learning between the two architectures in high uncertainty regions to further enhance performance. Our method achieves SOTA results on two public datasets and one private dataset.
Weakly Semi-supervised Whole Slide Image Classification by Two-level Cross Consistency Supervision
Apr 16, 2025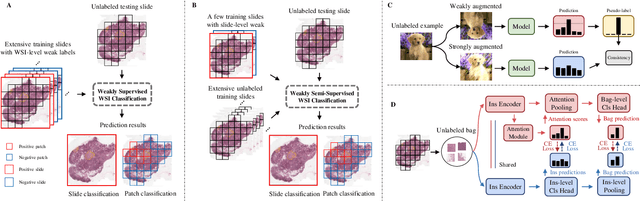
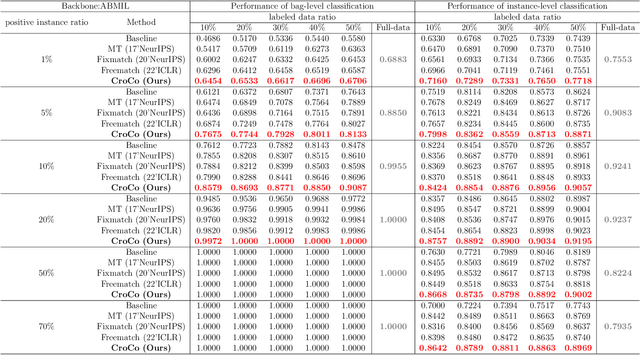
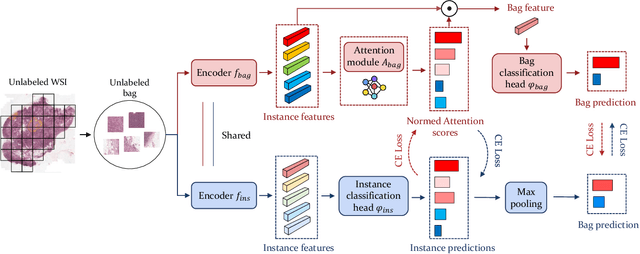
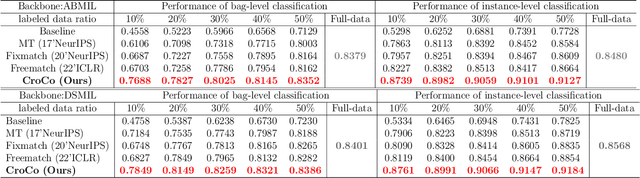
Abstract:Computer-aided Whole Slide Image (WSI) classification has the potential to enhance the accuracy and efficiency of clinical pathological diagnosis. It is commonly formulated as a Multiple Instance Learning (MIL) problem, where each WSI is treated as a bag and the small patches extracted from the WSI are considered instances within that bag. However, obtaining labels for a large number of bags is a costly and time-consuming process, particularly when utilizing existing WSIs for new classification tasks. This limitation renders most existing WSI classification methods ineffective. To address this issue, we propose a novel WSI classification problem setting, more aligned with clinical practice, termed Weakly Semi-supervised Whole slide image Classification (WSWC). In WSWC, a small number of bags are labeled, while a significant number of bags remain unlabeled. The MIL nature of the WSWC problem, coupled with the absence of patch labels, distinguishes it from typical semi-supervised image classification problems, making existing algorithms for natural images unsuitable for directly solving the WSWC problem. In this paper, we present a concise and efficient framework, named CroCo, to tackle the WSWC problem through two-level Cross Consistency supervision. CroCo comprises two heterogeneous classifier branches capable of performing both instance classification and bag classification. The fundamental idea is to establish cross-consistency supervision at both the bag-level and instance-level between the two branches during training. Extensive experiments conducted on four datasets demonstrate that CroCo achieves superior bag classification and instance classification performance compared to other comparative methods when limited WSIs with bag labels are available. To the best of our knowledge, this paper presents for the first time the WSWC problem and gives a successful resolution.
Towards Unified Molecule-Enhanced Pathology Image Representation Learning via Integrating Spatial Transcriptomics
Dec 01, 2024



Abstract:Recent advancements in multimodal pre-training models have significantly advanced computational pathology. However, current approaches predominantly rely on visual-language models, which may impose limitations from a molecular perspective and lead to performance bottlenecks. Here, we introduce a Unified Molecule-enhanced Pathology Image REpresentationn Learning framework (UMPIRE). UMPIRE aims to leverage complementary information from gene expression profiles to guide the multimodal pre-training, enhancing the molecular awareness of pathology image representation learning. We demonstrate that this molecular perspective provides a robust, task-agnostic training signal for learning pathology image embeddings. Due to the scarcity of paired data, approximately 4 million entries of spatial transcriptomics gene expression were collected to train the gene encoder. By leveraging powerful pre-trained encoders, UMPIRE aligns the encoders across over 697K pathology image-gene expression pairs. The performance of UMPIRE is demonstrated across various molecular-related downstream tasks, including gene expression prediction, spot classification, and mutation state prediction in whole slide images. Our findings highlight the effectiveness of multimodal data integration and open new avenues for exploring computational pathology enhanced by molecular perspectives. The code and pre-trained weights are available at https://github.com/Hanminghao/UMPIRE.
Local Implicit Wavelet Transformer for Arbitrary-Scale Super-Resolution
Nov 10, 2024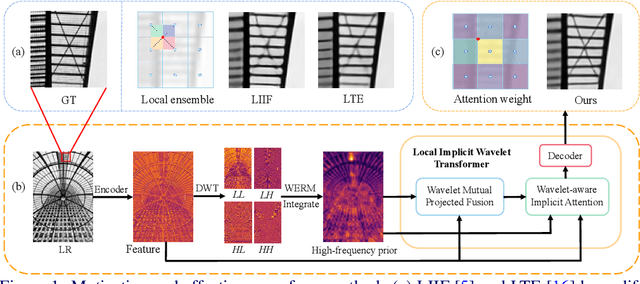
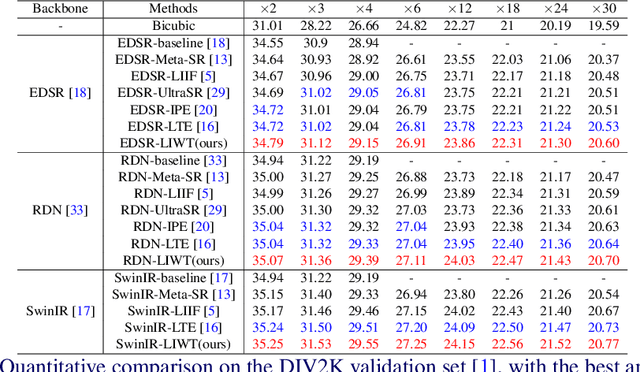

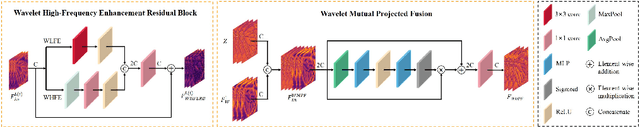
Abstract:Implicit neural representations have recently demonstrated promising potential in arbitrary-scale Super-Resolution (SR) of images. Most existing methods predict the pixel in the SR image based on the queried coordinate and ensemble nearby features, overlooking the importance of incorporating high-frequency prior information in images, which results in limited performance in reconstructing high-frequency texture details in images. To address this issue, we propose the Local Implicit Wavelet Transformer (LIWT) to enhance the restoration of high-frequency texture details. Specifically, we decompose the features extracted by an encoder into four sub-bands containing different frequency information using Discrete Wavelet Transform (DWT). We then introduce the Wavelet Enhanced Residual Module (WERM) to transform these four sub-bands into high-frequency priors, followed by utilizing the Wavelet Mutual Projected Fusion (WMPF) and the Wavelet-aware Implicit Attention (WIA) to fully exploit the high-frequency prior information for recovering high-frequency details in images. We conducted extensive experiments on benchmark datasets to validate the effectiveness of LIWT. Both qualitative and quantitative results demonstrate that LIWT achieves promising performance in arbitrary-scale SR tasks, outperforming other state-of-the-art methods. The code is available at https://github.com/dmhdmhdmh/LIWT.
FANCL: Feature-Guided Attention Network with Curriculum Learning for Brain Metastases Segmentation
Oct 29, 2024Abstract:Accurate segmentation of brain metastases (BMs) in MR image is crucial for the diagnosis and follow-up of patients. Methods based on deep convolutional neural networks (CNNs) have achieved high segmentation performance. However, due to the loss of critical feature information caused by convolutional and pooling operations, CNNs still face great challenges in small BMs segmentation. Besides, BMs are irregular and easily confused with healthy tissues, which makes it difficult for the model to effectively learn tumor structure during training. To address these issues, this paper proposes a novel model called feature-guided attention network with curriculum learning (FANCL). Based on CNNs, FANCL utilizes the input image and its feature to establish the intrinsic connections between metastases of different sizes, which can effectively compensate for the loss of high-level feature from small tumors with the information of large tumors. Furthermore, FANCL applies the voxel-level curriculum learning strategy to help the model gradually learn the structure and details of BMs. And baseline models of varying depths are employed as curriculum-mining networks for organizing the curriculum progression. The evaluation results on the BraTS-METS 2023 dataset indicate that FANCL significantly improves the segmentation performance, confirming the effectiveness of our method.
FAST: A Dual-tier Few-Shot Learning Paradigm for Whole Slide Image Classification
Sep 29, 2024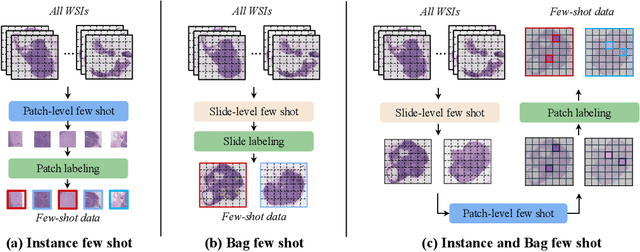
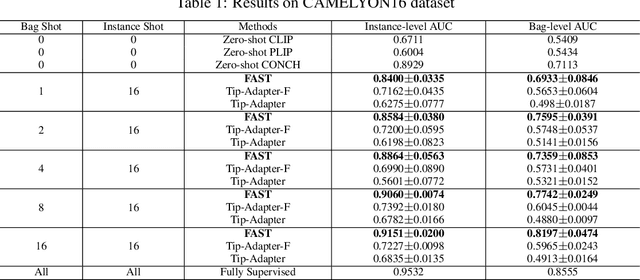
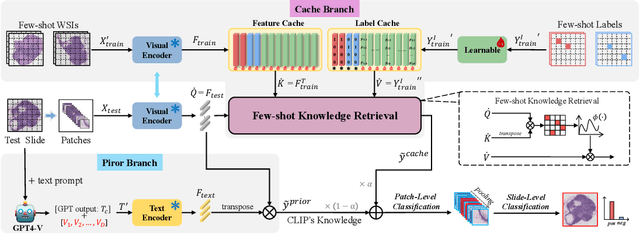
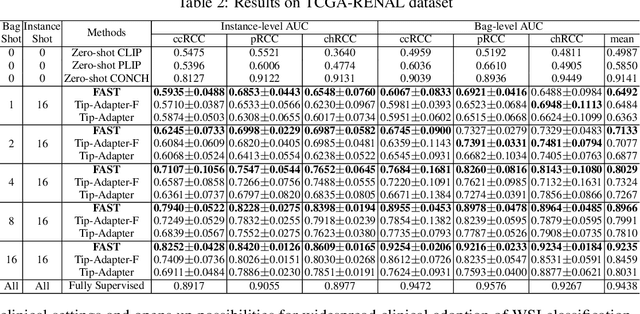
Abstract:The expensive fine-grained annotation and data scarcity have become the primary obstacles for the widespread adoption of deep learning-based Whole Slide Images (WSI) classification algorithms in clinical practice. Unlike few-shot learning methods in natural images that can leverage the labels of each image, existing few-shot WSI classification methods only utilize a small number of fine-grained labels or weakly supervised slide labels for training in order to avoid expensive fine-grained annotation. They lack sufficient mining of available WSIs, severely limiting WSI classification performance. To address the above issues, we propose a novel and efficient dual-tier few-shot learning paradigm for WSI classification, named FAST. FAST consists of a dual-level annotation strategy and a dual-branch classification framework. Firstly, to avoid expensive fine-grained annotation, we collect a very small number of WSIs at the slide level, and annotate an extremely small number of patches. Then, to fully mining the available WSIs, we use all the patches and available patch labels to build a cache branch, which utilizes the labeled patches to learn the labels of unlabeled patches and through knowledge retrieval for patch classification. In addition to the cache branch, we also construct a prior branch that includes learnable prompt vectors, using the text encoder of visual-language models for patch classification. Finally, we integrate the results from both branches to achieve WSI classification. Extensive experiments on binary and multi-class datasets demonstrate that our proposed method significantly surpasses existing few-shot classification methods and approaches the accuracy of fully supervised methods with only 0.22$\%$ annotation costs. All codes and models will be publicly available on https://github.com/fukexue/FAST.
MSCPT: Few-shot Whole Slide Image Classification with Multi-scale and Context-focused Prompt Tuning
Aug 21, 2024



Abstract:Multiple instance learning (MIL) has become a standard paradigm for weakly supervised classification of whole slide images (WSI). However, this paradigm relies on the use of a large number of labelled WSIs for training. The lack of training data and the presence of rare diseases present significant challenges for these methods. Prompt tuning combined with the pre-trained Vision-Language models (VLMs) is an effective solution to the Few-shot Weakly Supervised WSI classification (FSWC) tasks. Nevertheless, applying prompt tuning methods designed for natural images to WSIs presents three significant challenges: 1) These methods fail to fully leverage the prior knowledge from the VLM's text modality; 2) They overlook the essential multi-scale and contextual information in WSIs, leading to suboptimal results; and 3) They lack exploration of instance aggregation methods. To address these problems, we propose a Multi-Scale and Context-focused Prompt Tuning (MSCPT) method for FSWC tasks. Specifically, MSCPT employs the frozen large language model to generate pathological visual language prior knowledge at multi-scale, guiding hierarchical prompt tuning. Additionally, we design a graph prompt tuning module to learn essential contextual information within WSI, and finally, a non-parametric cross-guided instance aggregation module has been introduced to get the WSI-level features. Based on two VLMs, extensive experiments and visualizations on three datasets demonstrated the powerful performance of our MSCPT.
Advancing H&E-to-IHC Stain Translation in Breast Cancer: A Multi-Magnification and Attention-Based Approach
Aug 04, 2024
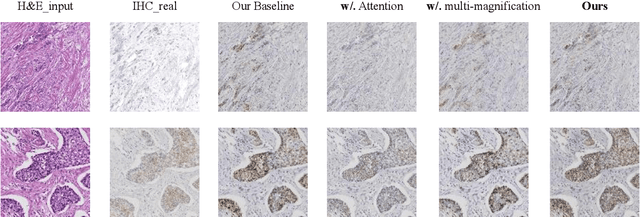


Abstract:Breast cancer presents a significant healthcare challenge globally, demanding precise diagnostics and effective treatment strategies, where histopathological examination of Hematoxylin and Eosin (H&E) stained tissue sections plays a central role. Despite its importance, evaluating specific biomarkers like Human Epidermal Growth Factor Receptor 2 (HER2) for personalized treatment remains constrained by the resource-intensive nature of Immunohistochemistry (IHC). Recent strides in deep learning, particularly in image-to-image translation, offer promise in synthesizing IHC-HER2 slides from H\&E stained slides. However, existing methodologies encounter challenges, including managing multiple magnifications in pathology images and insufficient focus on crucial information during translation. To address these issues, we propose a novel model integrating attention mechanisms and multi-magnification information processing. Our model employs a multi-magnification processing strategy to extract and utilize information from various magnifications within pathology images, facilitating robust image translation. Additionally, an attention module within the generative network prioritizes critical information for image distribution translation while minimizing less pertinent details. Rigorous testing on a publicly available breast cancer dataset demonstrates superior performance compared to existing methods, establishing our model as a state-of-the-art solution in advancing pathology image translation from H&E to IHC staining.
Multi-modal Data Binding for Survival Analysis Modeling with Incomplete Data and Annotations
Jul 25, 2024Abstract:Survival analysis stands as a pivotal process in cancer treatment research, crucial for predicting patient survival rates accurately. Recent advancements in data collection techniques have paved the way for enhancing survival predictions by integrating information from multiple modalities. However, real-world scenarios often present challenges with incomplete data, particularly when dealing with censored survival labels. Prior works have addressed missing modalities but have overlooked incomplete labels, which can introduce bias and limit model efficacy. To bridge this gap, we introduce a novel framework that simultaneously handles incomplete data across modalities and censored survival labels. Our approach employs advanced foundation models to encode individual modalities and align them into a universal representation space for seamless fusion. By generating pseudo labels and incorporating uncertainty, we significantly enhance predictive accuracy. The proposed method demonstrates outstanding prediction accuracy in two survival analysis tasks on both employed datasets. This innovative approach overcomes limitations associated with disparate modalities and improves the feasibility of comprehensive survival analysis using multiple large foundation models.
Deep Mutual Learning among Partially Labeled Datasets for Multi-Organ Segmentation
Jul 17, 2024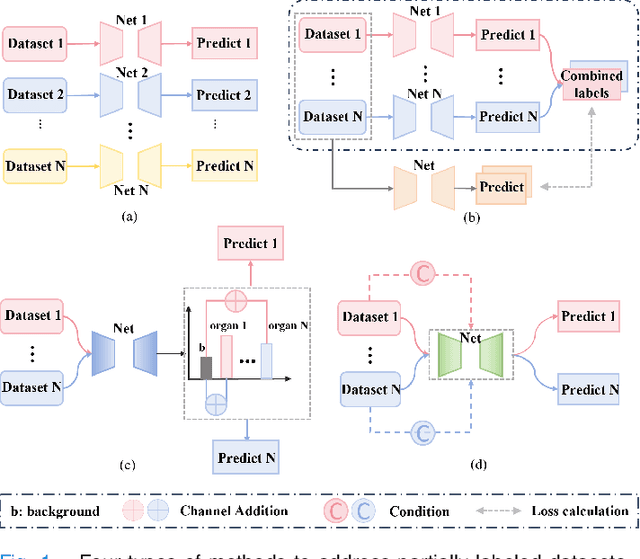
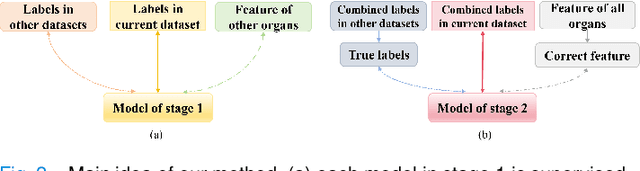
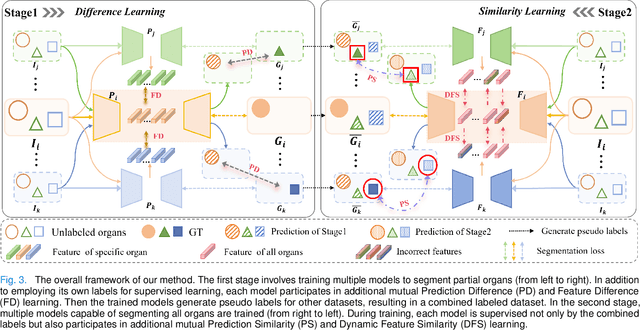
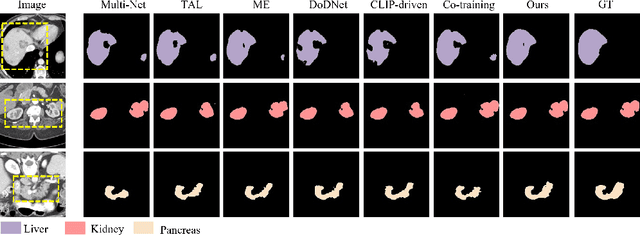
Abstract:The task of labeling multiple organs for segmentation is a complex and time-consuming process, resulting in a scarcity of comprehensively labeled multi-organ datasets while the emergence of numerous partially labeled datasets. Current methods are inadequate in effectively utilizing the supervised information available from these datasets, thereby impeding the progress in improving the segmentation accuracy. This paper proposes a two-stage multi-organ segmentation method based on mutual learning, aiming to improve multi-organ segmentation performance by complementing information among partially labeled datasets. In the first stage, each partial-organ segmentation model utilizes the non-overlapping organ labels from different datasets and the distinct organ features extracted by different models, introducing additional mutual difference learning to generate higher quality pseudo labels for unlabeled organs. In the second stage, each full-organ segmentation model is supervised by fully labeled datasets with pseudo labels and leverages true labels from other datasets, while dynamically sharing accurate features across different models, introducing additional mutual similarity learning to enhance multi-organ segmentation performance. Extensive experiments were conducted on nine datasets that included the head and neck, chest, abdomen, and pelvis. The results indicate that our method has achieved SOTA performance in segmentation tasks that rely on partial labels, and the ablation studies have thoroughly confirmed the efficacy of the mutual learning mechanism.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge