Shiman Li
Weakly Semi-supervised Whole Slide Image Classification by Two-level Cross Consistency Supervision
Apr 16, 2025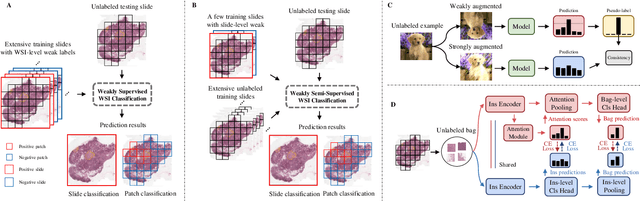
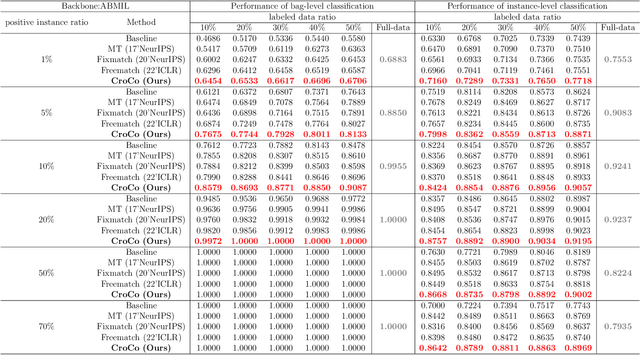
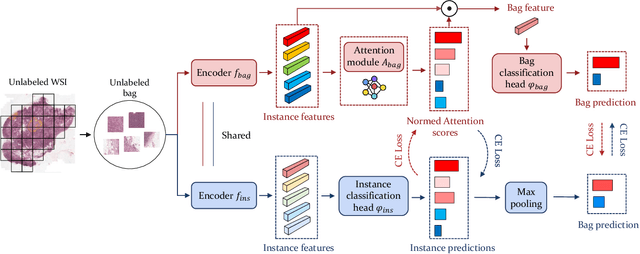
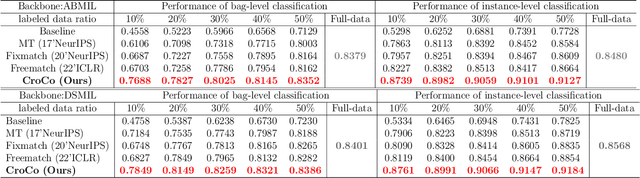
Abstract:Computer-aided Whole Slide Image (WSI) classification has the potential to enhance the accuracy and efficiency of clinical pathological diagnosis. It is commonly formulated as a Multiple Instance Learning (MIL) problem, where each WSI is treated as a bag and the small patches extracted from the WSI are considered instances within that bag. However, obtaining labels for a large number of bags is a costly and time-consuming process, particularly when utilizing existing WSIs for new classification tasks. This limitation renders most existing WSI classification methods ineffective. To address this issue, we propose a novel WSI classification problem setting, more aligned with clinical practice, termed Weakly Semi-supervised Whole slide image Classification (WSWC). In WSWC, a small number of bags are labeled, while a significant number of bags remain unlabeled. The MIL nature of the WSWC problem, coupled with the absence of patch labels, distinguishes it from typical semi-supervised image classification problems, making existing algorithms for natural images unsuitable for directly solving the WSWC problem. In this paper, we present a concise and efficient framework, named CroCo, to tackle the WSWC problem through two-level Cross Consistency supervision. CroCo comprises two heterogeneous classifier branches capable of performing both instance classification and bag classification. The fundamental idea is to establish cross-consistency supervision at both the bag-level and instance-level between the two branches during training. Extensive experiments conducted on four datasets demonstrate that CroCo achieves superior bag classification and instance classification performance compared to other comparative methods when limited WSIs with bag labels are available. To the best of our knowledge, this paper presents for the first time the WSWC problem and gives a successful resolution.
SP${ }^3$ : Superpixel-propagated pseudo-label learning for weakly semi-supervised medical image segmentation
Nov 18, 2024Abstract:Deep learning-based medical image segmentation helps assist diagnosis and accelerate the treatment process while the model training usually requires large-scale dense annotation datasets. Weakly semi-supervised medical image segmentation is an essential application because it only requires a small amount of scribbles and a large number of unlabeled data to train the model, which greatly reduces the clinician's effort to fully annotate images. To handle the inadequate supervisory information challenge in weakly semi-supervised segmentation (WSSS), a SuperPixel-Propagated Pseudo-label (SP${}^3$) learning method is proposed, using the structural information contained in superpixel for supplemental information. Specifically, the annotation of scribbles is propagated to superpixels and thus obtains a dense annotation for supervised training. Since the quality of pseudo-labels is limited by the low-quality annotation, the beneficial superpixels selected by dynamic thresholding are used to refine pseudo-labels. Furthermore, aiming to alleviate the negative impact of noise in pseudo-label, superpixel-level uncertainty is incorporated to guide the pseudo-label supervision for stable learning. Our method achieves state-of-the-art performance on both tumor and organ segmentation datasets under the WSSS setting, using only 3\% of the annotation workload compared to fully supervised methods and attaining approximately 80\% Dice score. Additionally, our method outperforms eight weakly and semi-supervised methods under both weakly supervised and semi-supervised settings. Results of extensive experiments validate the effectiveness and annotation efficiency of our weakly semi-supervised segmentation, which can assist clinicians in achieving automated segmentation for organs or tumors quickly and ultimately benefit patients.
MCSD: An Efficient Language Model with Diverse Fusion
Jun 18, 2024
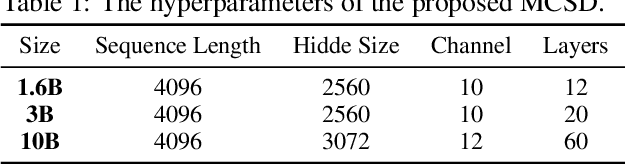

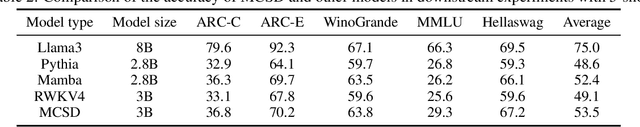
Abstract:Transformers excel in Natural Language Processing (NLP) due to their prowess in capturing long-term dependencies but suffer from exponential resource consumption with increasing sequence lengths. To address these challenges, we propose MCSD model, an efficient language model with linear scaling and fast inference speed. MCSD model leverages diverse feature fusion, primarily through the multi-channel slope and decay (MCSD) block, to robustly represent features. This block comprises slope and decay sections that extract features across diverse temporal receptive fields, facilitating capture of both local and global information. In addition, MCSD block conducts element-wise fusion of diverse features to further enhance the delicate feature extraction capability. For inference, we formulate the inference process into a recurrent representation, slashing space complexity to $O(1)$ and time complexity to $O(N)$ respectively. Our experiments show that MCSD attains higher throughput and lower GPU memory consumption compared to Transformers, while maintaining comparable performance to larger-scale language learning models on benchmark tests. These attributes position MCSD as a promising base for edge deployment and embodied intelligence.
A comprehensive survey on deep active learning and its applications in medical image analysis
Oct 24, 2023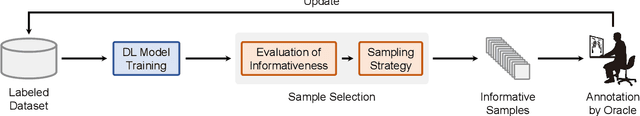
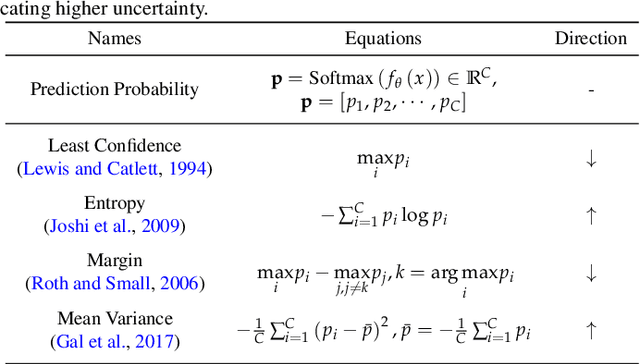
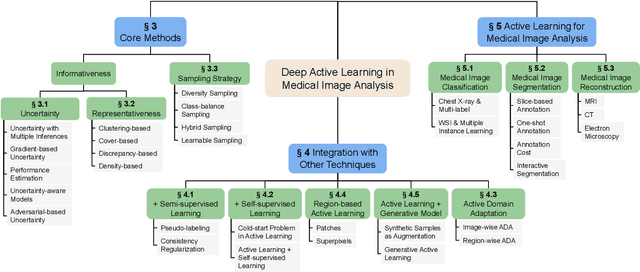
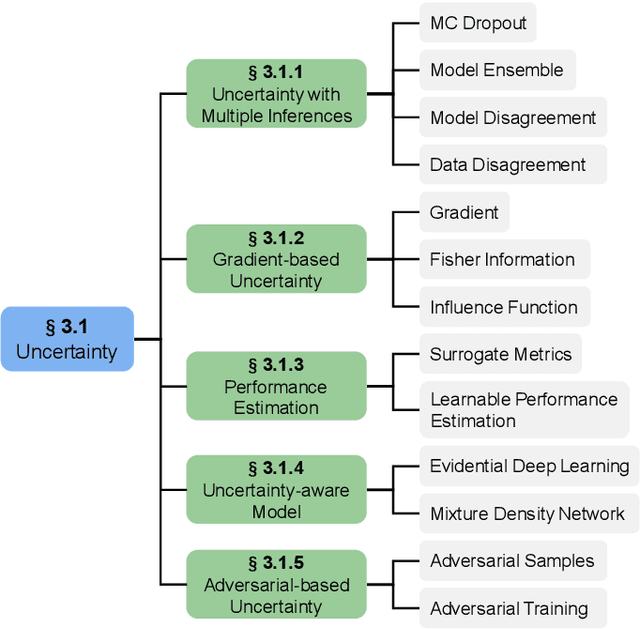
Abstract:Deep learning has achieved widespread success in medical image analysis, leading to an increasing demand for large-scale expert-annotated medical image datasets. Yet, the high cost of annotating medical images severely hampers the development of deep learning in this field. To reduce annotation costs, active learning aims to select the most informative samples for annotation and train high-performance models with as few labeled samples as possible. In this survey, we review the core methods of active learning, including the evaluation of informativeness and sampling strategy. For the first time, we provide a detailed summary of the integration of active learning with other label-efficient techniques, such as semi-supervised, self-supervised learning, and so on. Additionally, we also highlight active learning works that are specifically tailored to medical image analysis. In the end, we offer our perspectives on the future trends and challenges of active learning and its applications in medical image analysis.
Multi-organ segmentation: a progressive exploration of learning paradigms under scarce annotation
Feb 09, 2023Abstract:Precise delineation of multiple organs or abnormal regions in the human body from medical images plays an essential role in computer-aided diagnosis, surgical simulation, image-guided interventions, and especially in radiotherapy treatment planning. Thus, it is of great significance to explore automatic segmentation approaches, among which deep learning-based approaches have evolved rapidly and witnessed remarkable progress in multi-organ segmentation. However, obtaining an appropriately sized and fine-grained annotated dataset of multiple organs is extremely hard and expensive. Such scarce annotation limits the development of high-performance multi-organ segmentation models but promotes many annotation-efficient learning paradigms. Among these, studies on transfer learning leveraging external datasets, semi-supervised learning using unannotated datasets and partially-supervised learning integrating partially-labeled datasets have led the dominant way to break such dilemma in multi-organ segmentation. We first review the traditional fully supervised method, then present a comprehensive and systematic elaboration of the 3 abovementioned learning paradigms in the context of multi-organ segmentation from both technical and methodological perspectives, and finally summarize their challenges and future trends.
TransFuse: A Unified Transformer-based Image Fusion Framework using Self-supervised Learning
Jan 19, 2022Abstract:Image fusion is a technique to integrate information from multiple source images with complementary information to improve the richness of a single image. Due to insufficient task-specific training data and corresponding ground truth, most existing end-to-end image fusion methods easily fall into overfitting or tedious parameter optimization processes. Two-stage methods avoid the need of large amount of task-specific training data by training encoder-decoder network on large natural image datasets and utilizing the extracted features for fusion, but the domain gap between natural images and different fusion tasks results in limited performance. In this study, we design a novel encoder-decoder based image fusion framework and propose a destruction-reconstruction based self-supervised training scheme to encourage the network to learn task-specific features. Specifically, we propose three destruction-reconstruction self-supervised auxiliary tasks for multi-modal image fusion, multi-exposure image fusion and multi-focus image fusion based on pixel intensity non-linear transformation, brightness transformation and noise transformation, respectively. In order to encourage different fusion tasks to promote each other and increase the generalizability of the trained network, we integrate the three self-supervised auxiliary tasks by randomly choosing one of them to destroy a natural image in model training. In addition, we design a new encoder that combines CNN and Transformer for feature extraction, so that the trained model can exploit both local and global information. Extensive experiments on multi-modal image fusion, multi-exposure image fusion and multi-focus image fusion tasks demonstrate that our proposed method achieves the state-of-the-art performance in both subjective and objective evaluations. The code will be publicly available soon.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge