Jiayue Zhao
GLM-5: from Vibe Coding to Agentic Engineering
Feb 17, 2026Abstract:We present GLM-5, a next-generation foundation model designed to transition the paradigm of vibe coding to agentic engineering. Building upon the agentic, reasoning, and coding (ARC) capabilities of its predecessor, GLM-5 adopts DSA to significantly reduce training and inference costs while maintaining long-context fidelity. To advance model alignment and autonomy, we implement a new asynchronous reinforcement learning infrastructure that drastically improves post-training efficiency by decoupling generation from training. Furthermore, we propose novel asynchronous agent RL algorithms that further improve RL quality, enabling the model to learn from complex, long-horizon interactions more effectively. Through these innovations, GLM-5 achieves state-of-the-art performance on major open benchmarks. Most critically, GLM-5 demonstrates unprecedented capability in real-world coding tasks, surpassing previous baselines in handling end-to-end software engineering challenges. Code, models, and more information are available at https://github.com/zai-org/GLM-5.
View-on-Graph: Zero-shot 3D Visual Grounding via Vision-Language Reasoning on Scene Graphs
Dec 10, 2025Abstract:3D visual grounding (3DVG) identifies objects in 3D scenes from language descriptions. Existing zero-shot approaches leverage 2D vision-language models (VLMs) by converting 3D spatial information (SI) into forms amenable to VLM processing, typically as composite inputs such as specified view renderings or video sequences with overlaid object markers. However, this VLM + SI paradigm yields entangled visual representations that compel the VLM to process entire cluttered cues, making it hard to exploit spatial semantic relationships effectively. In this work, we propose a new VLM x SI paradigm that externalizes the 3D SI into a form enabling the VLM to incrementally retrieve only what it needs during reasoning. We instantiate this paradigm with a novel View-on-Graph (VoG) method, which organizes the scene into a multi-modal, multi-layer scene graph and allows the VLM to operate as an active agent that selectively accesses necessary cues as it traverses the scene. This design offers two intrinsic advantages: (i) by structuring 3D context into a spatially and semantically coherent scene graph rather than confounding the VLM with densely entangled visual inputs, it lowers the VLM's reasoning difficulty; and (ii) by actively exploring and reasoning over the scene graph, it naturally produces transparent, step-by-step traces for interpretable 3DVG. Extensive experiments show that VoG achieves state-of-the-art zero-shot performance, establishing structured scene exploration as a promising strategy for advancing zero-shot 3DVG.
GLM-4.5: Agentic, Reasoning, and Coding (ARC) Foundation Models
Aug 08, 2025Abstract:We present GLM-4.5, an open-source Mixture-of-Experts (MoE) large language model with 355B total parameters and 32B activated parameters, featuring a hybrid reasoning method that supports both thinking and direct response modes. Through multi-stage training on 23T tokens and comprehensive post-training with expert model iteration and reinforcement learning, GLM-4.5 achieves strong performance across agentic, reasoning, and coding (ARC) tasks, scoring 70.1% on TAU-Bench, 91.0% on AIME 24, and 64.2% on SWE-bench Verified. With much fewer parameters than several competitors, GLM-4.5 ranks 3rd overall among all evaluated models and 2nd on agentic benchmarks. We release both GLM-4.5 (355B parameters) and a compact version, GLM-4.5-Air (106B parameters), to advance research in reasoning and agentic AI systems. Code, models, and more information are available at https://github.com/zai-org/GLM-4.5.
SP${ }^3$ : Superpixel-propagated pseudo-label learning for weakly semi-supervised medical image segmentation
Nov 18, 2024Abstract:Deep learning-based medical image segmentation helps assist diagnosis and accelerate the treatment process while the model training usually requires large-scale dense annotation datasets. Weakly semi-supervised medical image segmentation is an essential application because it only requires a small amount of scribbles and a large number of unlabeled data to train the model, which greatly reduces the clinician's effort to fully annotate images. To handle the inadequate supervisory information challenge in weakly semi-supervised segmentation (WSSS), a SuperPixel-Propagated Pseudo-label (SP${}^3$) learning method is proposed, using the structural information contained in superpixel for supplemental information. Specifically, the annotation of scribbles is propagated to superpixels and thus obtains a dense annotation for supervised training. Since the quality of pseudo-labels is limited by the low-quality annotation, the beneficial superpixels selected by dynamic thresholding are used to refine pseudo-labels. Furthermore, aiming to alleviate the negative impact of noise in pseudo-label, superpixel-level uncertainty is incorporated to guide the pseudo-label supervision for stable learning. Our method achieves state-of-the-art performance on both tumor and organ segmentation datasets under the WSSS setting, using only 3\% of the annotation workload compared to fully supervised methods and attaining approximately 80\% Dice score. Additionally, our method outperforms eight weakly and semi-supervised methods under both weakly supervised and semi-supervised settings. Results of extensive experiments validate the effectiveness and annotation efficiency of our weakly semi-supervised segmentation, which can assist clinicians in achieving automated segmentation for organs or tumors quickly and ultimately benefit patients.
Towards more precise automatic analysis: a comprehensive survey of deep learning-based multi-organ segmentation
Mar 02, 2023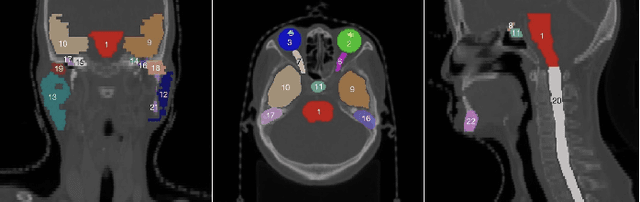
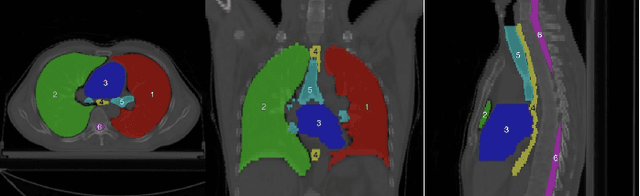
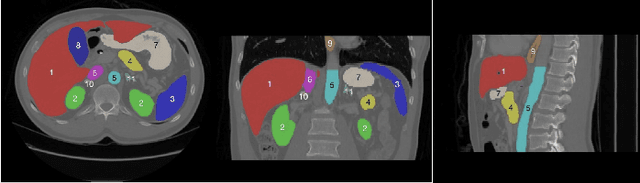
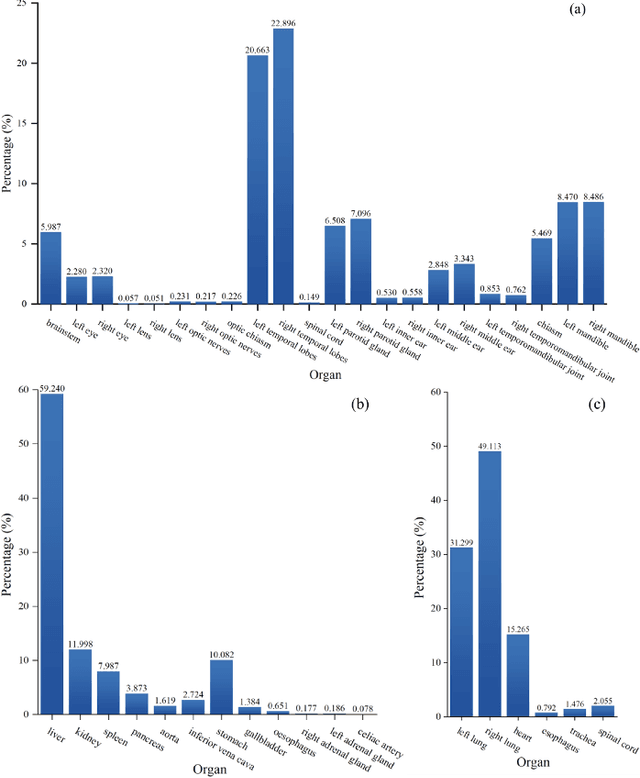
Abstract:Accurate segmentation of multiple organs of the head, neck, chest, and abdomen from medical images is an essential step in computer-aided diagnosis, surgical navigation, and radiation therapy. In the past few years, with a data-driven feature extraction approach and end-to-end training, automatic deep learning-based multi-organ segmentation method has far outperformed traditional methods and become a new research topic. This review systematically summarizes the latest research in this field. For the first time, from the perspective of full and imperfect annotation, we comprehensively compile 161 studies on deep learning-based multi-organ segmentation in multiple regions such as the head and neck, chest, and abdomen, containing a total of 214 related references. The method based on full annotation summarizes the existing methods from four aspects: network architecture, network dimension, network dedicated modules, and network loss function. The method based on imperfect annotation summarizes the existing methods from two aspects: weak annotation-based methods and semi annotation-based methods. We also summarize frequently used datasets for multi-organ segmentation and discuss new challenges and new research trends in this field.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge