Yonghong Shi
HUR-MACL: High-Uncertainty Region-Guided Multi-Architecture Collaborative Learning for Head and Neck Multi-Organ Segmentation
Jan 08, 2026Abstract:Accurate segmentation of organs at risk in the head and neck is essential for radiation therapy, yet deep learning models often fail on small, complexly shaped organs. While hybrid architectures that combine different models show promise, they typically just concatenate features without exploiting the unique strengths of each component. This results in functional overlap and limited segmentation accuracy. To address these issues, we propose a high uncertainty region-guided multi-architecture collaborative learning (HUR-MACL) model for multi-organ segmentation in the head and neck. This model adaptively identifies high uncertainty regions using a convolutional neural network, and for these regions, Vision Mamba as well as Deformable CNN are utilized to jointly improve their segmentation accuracy. Additionally, a heterogeneous feature distillation loss was proposed to promote collaborative learning between the two architectures in high uncertainty regions to further enhance performance. Our method achieves SOTA results on two public datasets and one private dataset.
DM-Mamba: Dual-domain Multi-scale Mamba for MRI reconstruction
Jan 14, 2025Abstract:The accelerated MRI reconstruction poses a challenging ill-posed inverse problem due to the significant undersampling in k-space. Deep neural networks, such as CNNs and ViT, have shown substantial performance improvements for this task while encountering the dilemma between global receptive fields and efficient computation. To this end, this paper pioneers exploring Mamba, a new paradigm for long-range dependency modeling with linear complexity, for efficient and effective MRI reconstruction. However, directly applying Mamba to MRI reconstruction faces three significant issues: (1) Mamba's row-wise and column-wise scanning disrupts k-space's unique spectrum, leaving its potential in k-space learning unexplored. (2) Existing Mamba methods unfold feature maps with multiple lengthy scanning paths, leading to long-range forgetting and high computational burden. (3) Mamba struggles with spatially-varying contents, resulting in limited diversity of local representations. To address these, we propose a dual-domain multi-scale Mamba for MRI reconstruction from the following perspectives: (1) We pioneer vision Mamba in k-space learning. A circular scanning is customized for spectrum unfolding, benefiting the global modeling of k-space. (2) We propose a multi-scale Mamba with an efficient scanning strategy in both image and k-space domains. It mitigates long-range forgetting and achieves a better trade-off between efficiency and performance. (3) We develop a local diversity enhancement module to improve the spatially-varying representation of Mamba. Extensive experiments are conducted on three public datasets for MRI reconstruction under various undersampling patterns. Comprehensive results demonstrate that our method significantly outperforms state-of-the-art methods with lower computational cost. Implementation code will be available at https://github.com/XiaoMengLiLiLi/DM-Mamba.
Boosting ViT-based MRI Reconstruction from the Perspectives of Frequency Modulation, Spatial Purification, and Scale Diversification
Dec 14, 2024Abstract:The accelerated MRI reconstruction process presents a challenging ill-posed inverse problem due to the extensive under-sampling in k-space. Recently, Vision Transformers (ViTs) have become the mainstream for this task, demonstrating substantial performance improvements. However, there are still three significant issues remain unaddressed: (1) ViTs struggle to capture high-frequency components of images, limiting their ability to detect local textures and edge information, thereby impeding MRI restoration; (2) Previous methods calculate multi-head self-attention (MSA) among both related and unrelated tokens in content, introducing noise and significantly increasing computational burden; (3) The naive feed-forward network in ViTs cannot model the multi-scale information that is important for image restoration. In this paper, we propose FPS-Former, a powerful ViT-based framework, to address these issues from the perspectives of frequency modulation, spatial purification, and scale diversification. Specifically, for issue (1), we introduce a frequency modulation attention module to enhance the self-attention map by adaptively re-calibrating the frequency information in a Laplacian pyramid. For issue (2), we customize a spatial purification attention module to capture interactions among closely related tokens, thereby reducing redundant or irrelevant feature representations. For issue (3), we propose an efficient feed-forward network based on a hybrid-scale fusion strategy. Comprehensive experiments conducted on three public datasets show that our FPS-Former outperforms state-of-the-art methods while requiring lower computational costs.
Continuous K-space Recovery Network with Image Guidance for Fast MRI Reconstruction
Nov 18, 2024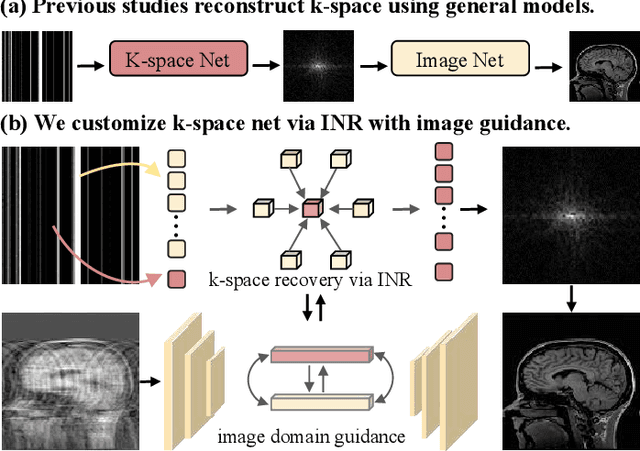
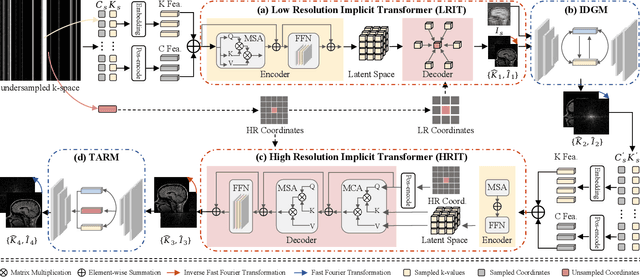
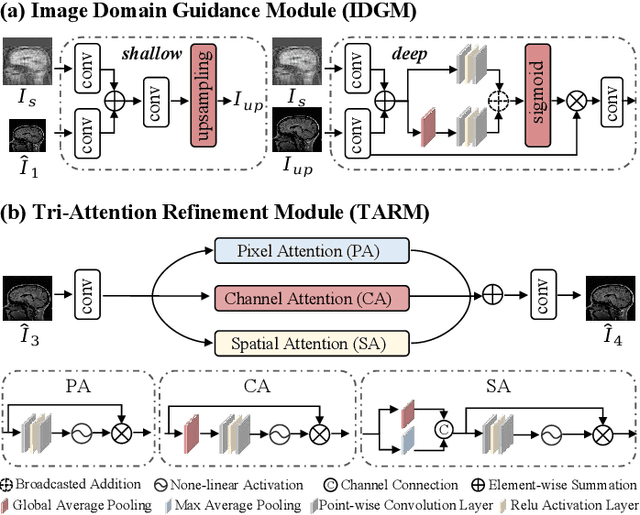
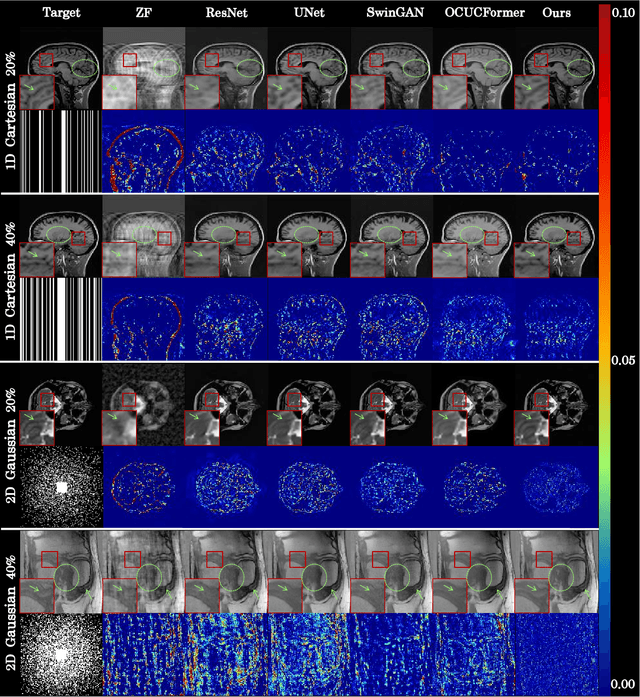
Abstract:Magnetic resonance imaging (MRI) is a crucial tool for clinical diagnosis while facing the challenge of long scanning time. To reduce the acquisition time, fast MRI reconstruction aims to restore high-quality images from the undersampled k-space. Existing methods typically train deep learning models to map the undersampled data to artifact-free MRI images. However, these studies often overlook the unique properties of k-space and directly apply general networks designed for image processing to k-space recovery, leaving the precise learning of k-space largely underexplored. In this work, we propose a continuous k-space recovery network from a new perspective of implicit neural representation with image domain guidance, which boosts the performance of MRI reconstruction. Specifically, (1) an implicit neural representation based encoder-decoder structure is customized to continuously query unsampled k-values. (2) an image guidance module is designed to mine the semantic information from the low-quality MRI images to further guide the k-space recovery. (3) a multi-stage training strategy is proposed to recover dense k-space progressively. Extensive experiments conducted on CC359, fastMRI, and IXI datasets demonstrate the effectiveness of our method and its superiority over other competitors.
FANCL: Feature-Guided Attention Network with Curriculum Learning for Brain Metastases Segmentation
Oct 29, 2024Abstract:Accurate segmentation of brain metastases (BMs) in MR image is crucial for the diagnosis and follow-up of patients. Methods based on deep convolutional neural networks (CNNs) have achieved high segmentation performance. However, due to the loss of critical feature information caused by convolutional and pooling operations, CNNs still face great challenges in small BMs segmentation. Besides, BMs are irregular and easily confused with healthy tissues, which makes it difficult for the model to effectively learn tumor structure during training. To address these issues, this paper proposes a novel model called feature-guided attention network with curriculum learning (FANCL). Based on CNNs, FANCL utilizes the input image and its feature to establish the intrinsic connections between metastases of different sizes, which can effectively compensate for the loss of high-level feature from small tumors with the information of large tumors. Furthermore, FANCL applies the voxel-level curriculum learning strategy to help the model gradually learn the structure and details of BMs. And baseline models of varying depths are employed as curriculum-mining networks for organizing the curriculum progression. The evaluation results on the BraTS-METS 2023 dataset indicate that FANCL significantly improves the segmentation performance, confirming the effectiveness of our method.
Deep Mutual Learning among Partially Labeled Datasets for Multi-Organ Segmentation
Jul 17, 2024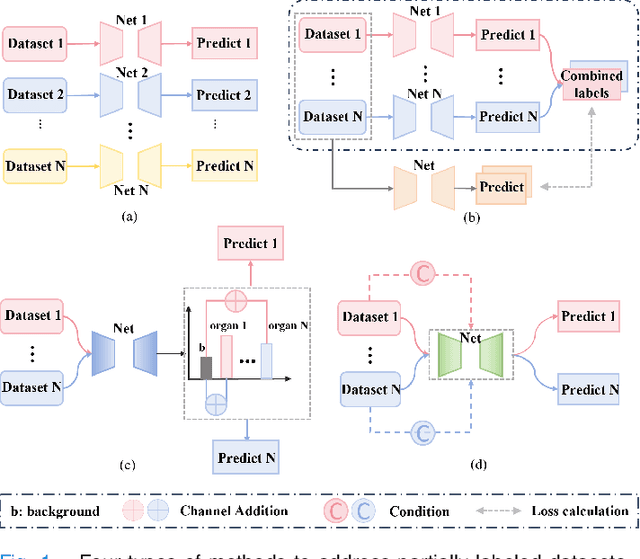
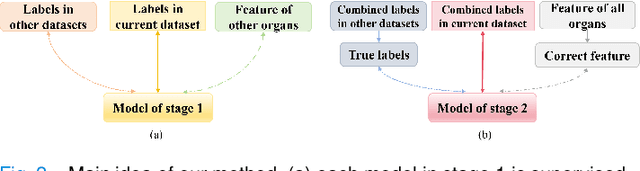
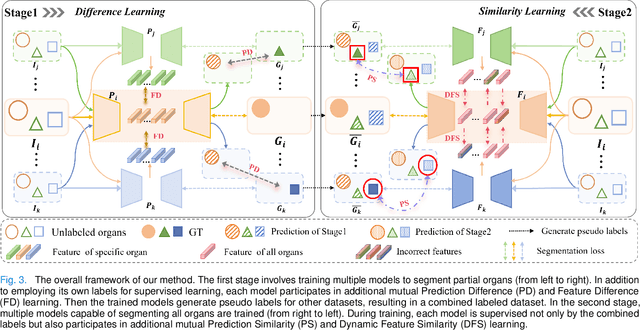
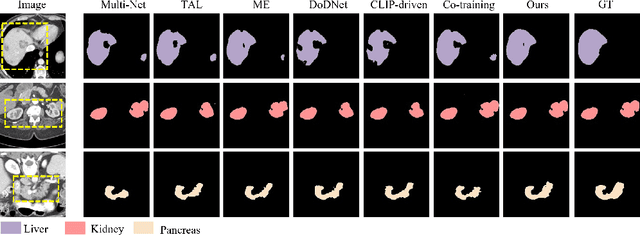
Abstract:The task of labeling multiple organs for segmentation is a complex and time-consuming process, resulting in a scarcity of comprehensively labeled multi-organ datasets while the emergence of numerous partially labeled datasets. Current methods are inadequate in effectively utilizing the supervised information available from these datasets, thereby impeding the progress in improving the segmentation accuracy. This paper proposes a two-stage multi-organ segmentation method based on mutual learning, aiming to improve multi-organ segmentation performance by complementing information among partially labeled datasets. In the first stage, each partial-organ segmentation model utilizes the non-overlapping organ labels from different datasets and the distinct organ features extracted by different models, introducing additional mutual difference learning to generate higher quality pseudo labels for unlabeled organs. In the second stage, each full-organ segmentation model is supervised by fully labeled datasets with pseudo labels and leverages true labels from other datasets, while dynamically sharing accurate features across different models, introducing additional mutual similarity learning to enhance multi-organ segmentation performance. Extensive experiments were conducted on nine datasets that included the head and neck, chest, abdomen, and pelvis. The results indicate that our method has achieved SOTA performance in segmentation tasks that rely on partial labels, and the ablation studies have thoroughly confirmed the efficacy of the mutual learning mechanism.
Towards more precise automatic analysis: a comprehensive survey of deep learning-based multi-organ segmentation
Mar 02, 2023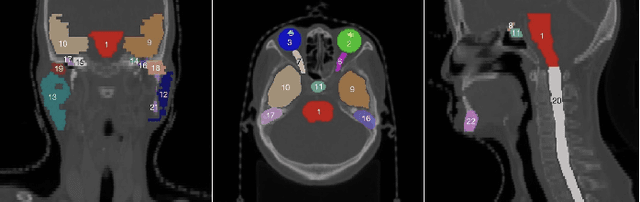
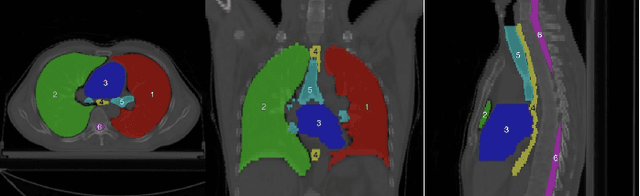
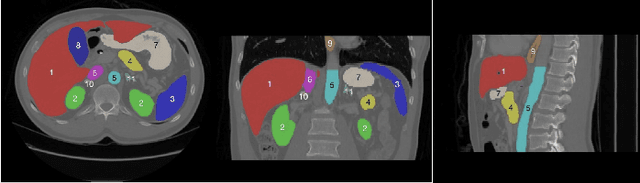
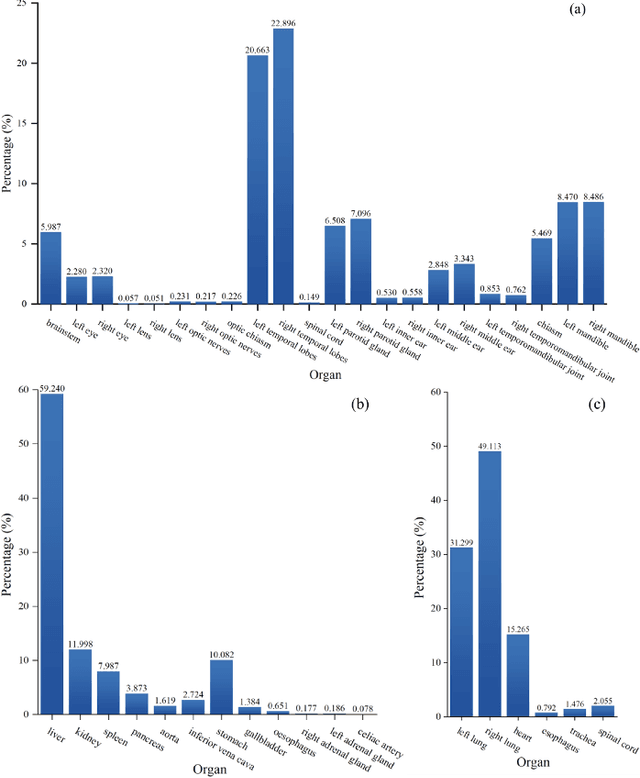
Abstract:Accurate segmentation of multiple organs of the head, neck, chest, and abdomen from medical images is an essential step in computer-aided diagnosis, surgical navigation, and radiation therapy. In the past few years, with a data-driven feature extraction approach and end-to-end training, automatic deep learning-based multi-organ segmentation method has far outperformed traditional methods and become a new research topic. This review systematically summarizes the latest research in this field. For the first time, from the perspective of full and imperfect annotation, we comprehensively compile 161 studies on deep learning-based multi-organ segmentation in multiple regions such as the head and neck, chest, and abdomen, containing a total of 214 related references. The method based on full annotation summarizes the existing methods from four aspects: network architecture, network dimension, network dedicated modules, and network loss function. The method based on imperfect annotation summarizes the existing methods from two aspects: weak annotation-based methods and semi annotation-based methods. We also summarize frequently used datasets for multi-organ segmentation and discuss new challenges and new research trends in this field.
Residual Block-based Multi-Label Classification and Localization Network with Integral Regression for Vertebrae Labeling
Jan 01, 2020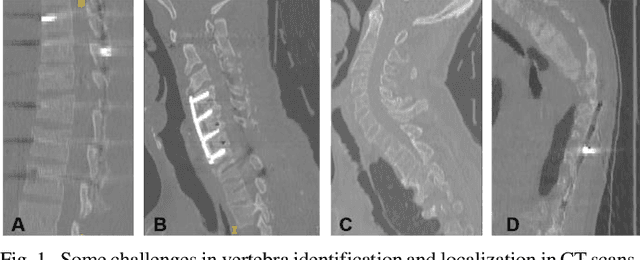
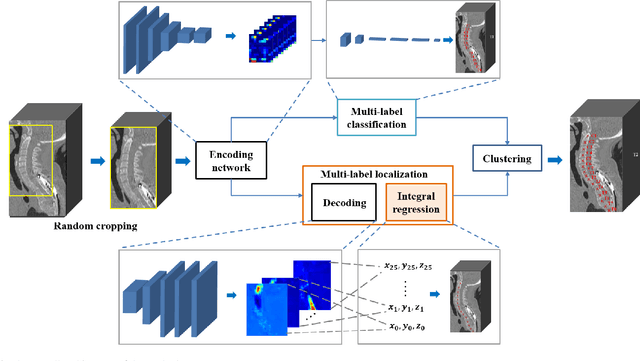
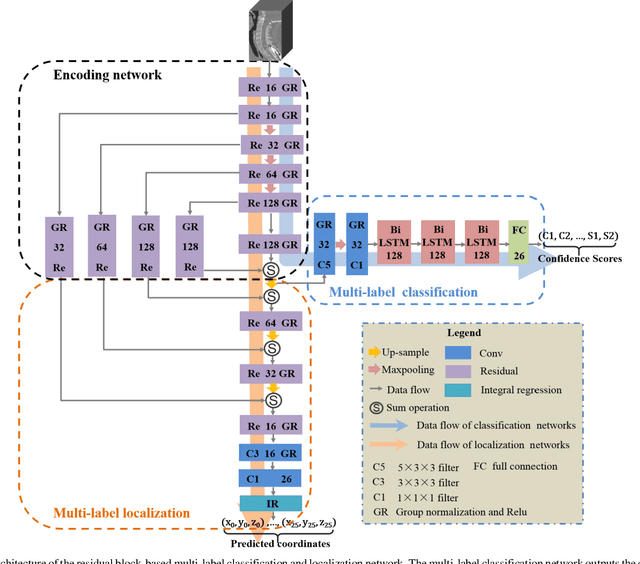
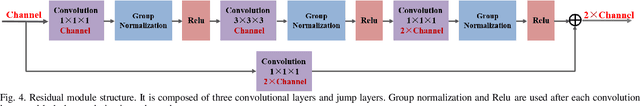
Abstract:Accurate identification and localization of the vertebrae in CT scans is a critical and standard preprocessing step for clinical spinal diagnosis and treatment. Existing methods are mainly based on the integration of multiple neural networks, and most of them use the Gaussian heat map to locate the vertebrae's centroid. However, the process of obtaining the vertebrae's centroid coordinates using heat maps is non-differentiable, so it is impossible to train the network to label the vertebrae directly. Therefore, for end-to-end differential training of vertebra coordinates on CT scans, a robust and accurate automatic vertebral labeling algorithm is proposed in this study. Firstly, a novel residual-based multi-label classification and localization network is developed, which can capture multi-scale features, but also utilize the residual module and skip connection to fuse the multi-level features. Secondly, to solve the problem that the process of finding coordinates is non-differentiable and the spatial structure is not destructible, integral regression module is used in the localization network. It combines the advantages of heat map representation and direct regression coordinates to achieve end-to-end training, and can be compatible with any key point detection methods of medical image based on heat map. Finally, multi-label classification of vertebrae is carried out, which use bidirectional long short term memory (Bi-LSTM) to enhance the learning of long contextual information to improve the classification performance. The proposed method is evaluated on a challenging dataset and the results are significantly better than the state-of-the-art methods (mean localization error <3mm).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge