Xiaoyuan Luo
Weakly Semi-supervised Whole Slide Image Classification by Two-level Cross Consistency Supervision
Apr 16, 2025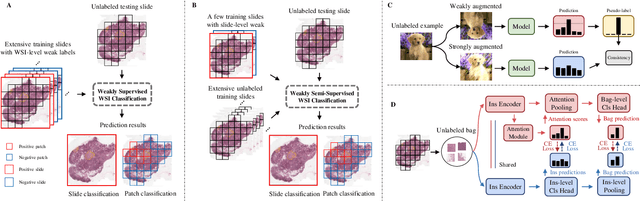
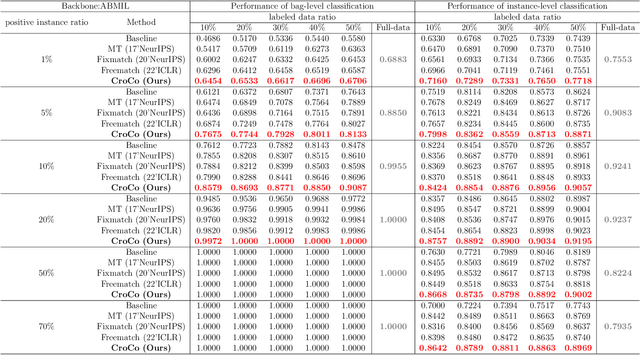
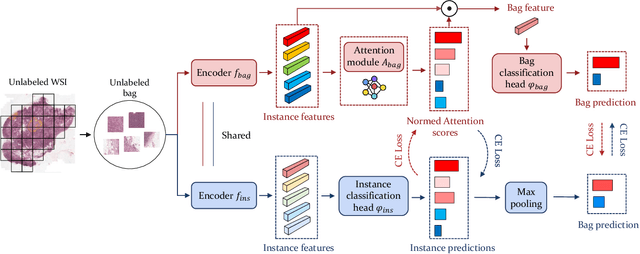
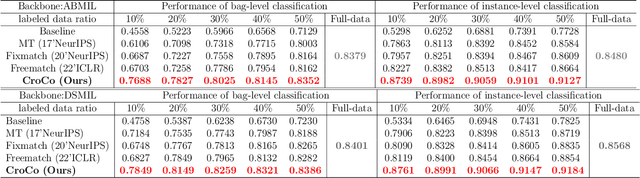
Abstract:Computer-aided Whole Slide Image (WSI) classification has the potential to enhance the accuracy and efficiency of clinical pathological diagnosis. It is commonly formulated as a Multiple Instance Learning (MIL) problem, where each WSI is treated as a bag and the small patches extracted from the WSI are considered instances within that bag. However, obtaining labels for a large number of bags is a costly and time-consuming process, particularly when utilizing existing WSIs for new classification tasks. This limitation renders most existing WSI classification methods ineffective. To address this issue, we propose a novel WSI classification problem setting, more aligned with clinical practice, termed Weakly Semi-supervised Whole slide image Classification (WSWC). In WSWC, a small number of bags are labeled, while a significant number of bags remain unlabeled. The MIL nature of the WSWC problem, coupled with the absence of patch labels, distinguishes it from typical semi-supervised image classification problems, making existing algorithms for natural images unsuitable for directly solving the WSWC problem. In this paper, we present a concise and efficient framework, named CroCo, to tackle the WSWC problem through two-level Cross Consistency supervision. CroCo comprises two heterogeneous classifier branches capable of performing both instance classification and bag classification. The fundamental idea is to establish cross-consistency supervision at both the bag-level and instance-level between the two branches during training. Extensive experiments conducted on four datasets demonstrate that CroCo achieves superior bag classification and instance classification performance compared to other comparative methods when limited WSIs with bag labels are available. To the best of our knowledge, this paper presents for the first time the WSWC problem and gives a successful resolution.
FAST: A Dual-tier Few-Shot Learning Paradigm for Whole Slide Image Classification
Sep 29, 2024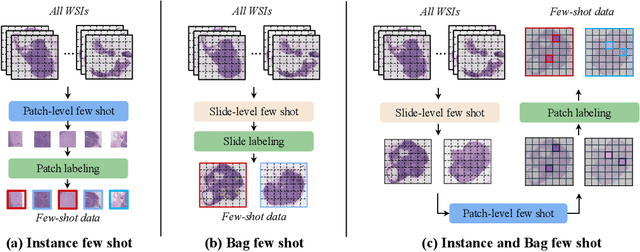
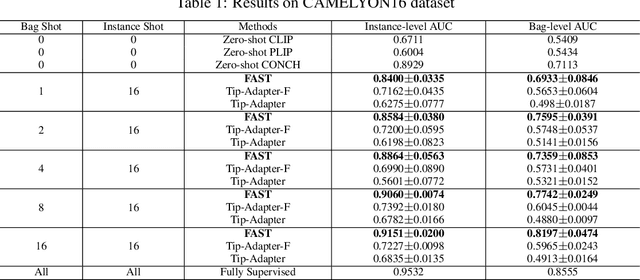
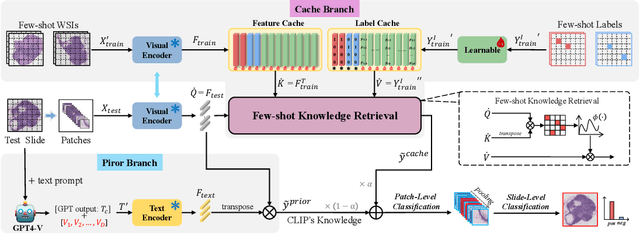
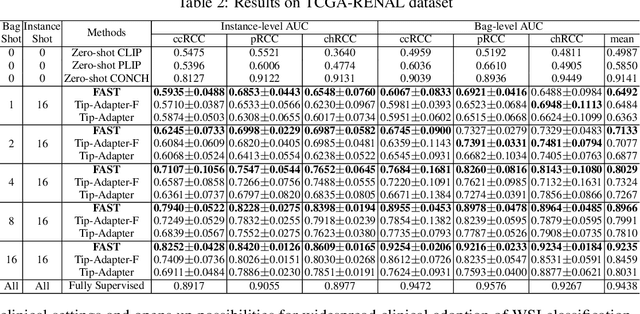
Abstract:The expensive fine-grained annotation and data scarcity have become the primary obstacles for the widespread adoption of deep learning-based Whole Slide Images (WSI) classification algorithms in clinical practice. Unlike few-shot learning methods in natural images that can leverage the labels of each image, existing few-shot WSI classification methods only utilize a small number of fine-grained labels or weakly supervised slide labels for training in order to avoid expensive fine-grained annotation. They lack sufficient mining of available WSIs, severely limiting WSI classification performance. To address the above issues, we propose a novel and efficient dual-tier few-shot learning paradigm for WSI classification, named FAST. FAST consists of a dual-level annotation strategy and a dual-branch classification framework. Firstly, to avoid expensive fine-grained annotation, we collect a very small number of WSIs at the slide level, and annotate an extremely small number of patches. Then, to fully mining the available WSIs, we use all the patches and available patch labels to build a cache branch, which utilizes the labeled patches to learn the labels of unlabeled patches and through knowledge retrieval for patch classification. In addition to the cache branch, we also construct a prior branch that includes learnable prompt vectors, using the text encoder of visual-language models for patch classification. Finally, we integrate the results from both branches to achieve WSI classification. Extensive experiments on binary and multi-class datasets demonstrate that our proposed method significantly surpasses existing few-shot classification methods and approaches the accuracy of fully supervised methods with only 0.22$\%$ annotation costs. All codes and models will be publicly available on https://github.com/fukexue/FAST.
Rethinking Multiple Instance Learning: Developing an Instance-Level Classifier via Weakly-Supervised Self-Training
Aug 09, 2024Abstract:Multiple instance learning (MIL) problem is currently solved from either bag-classification or instance-classification perspective, both of which ignore important information contained in some instances and result in limited performance. For example, existing methods often face difficulty in learning hard positive instances. In this paper, we formulate MIL as a semi-supervised instance classification problem, so that all the labeled and unlabeled instances can be fully utilized to train a better classifier. The difficulty in this formulation is that all the labeled instances are negative in MIL, and traditional self-training techniques used in semi-supervised learning tend to degenerate in generating pseudo labels for the unlabeled instances in this scenario. To resolve this problem, we propose a weakly-supervised self-training method, in which we utilize the positive bag labels to construct a global constraint and a local constraint on the pseudo labels to prevent them from degenerating and force the classifier to learn hard positive instances. It is worth noting that easy positive instances are instances are far from the decision boundary in the classification process, while hard positive instances are those close to the decision boundary. Through iterative optimization, the pseudo labels can gradually approach the true labels. Extensive experiments on two MNIST synthetic datasets, five traditional MIL benchmark datasets and two histopathology whole slide image datasets show that our method achieved new SOTA performance on all of them. The code will be publicly available.
Knowledge Extraction and Distillation from Large-Scale Image-Text Colonoscopy Records Leveraging Large Language and Vision Models
Oct 17, 2023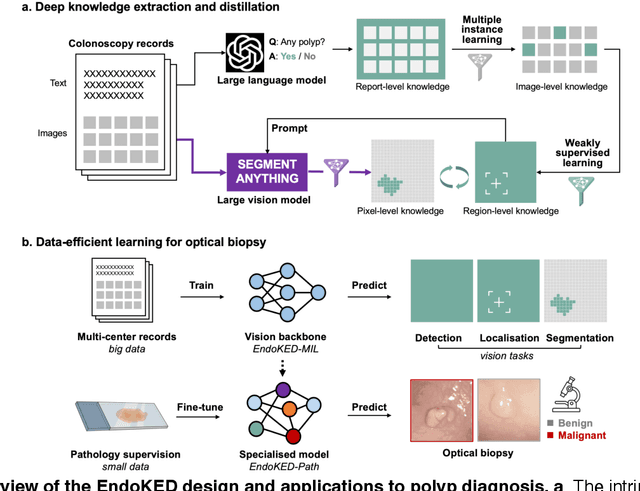
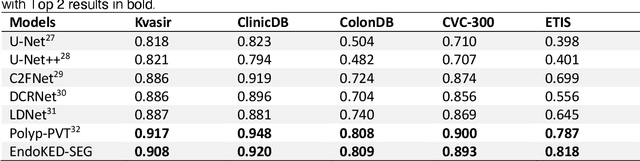
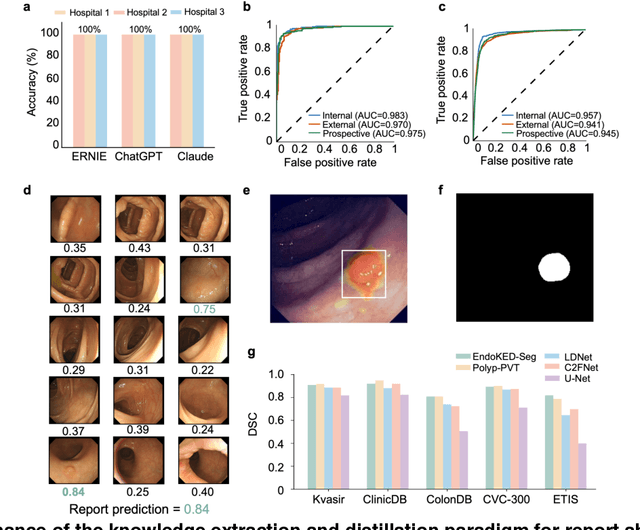
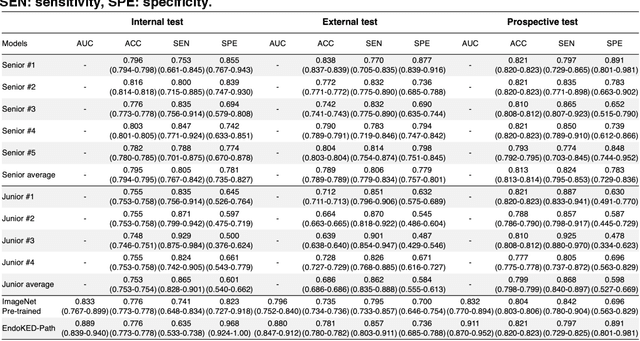
Abstract:The development of artificial intelligence systems for colonoscopy analysis often necessitates expert-annotated image datasets. However, limitations in dataset size and diversity impede model performance and generalisation. Image-text colonoscopy records from routine clinical practice, comprising millions of images and text reports, serve as a valuable data source, though annotating them is labour-intensive. Here we leverage recent advancements in large language and vision models and propose EndoKED, a data mining paradigm for deep knowledge extraction and distillation. EndoKED automates the transformation of raw colonoscopy records into image datasets with pixel-level annotation. We validate EndoKED using multi-centre datasets of raw colonoscopy records (~1 million images), demonstrating its superior performance in training polyp detection and segmentation models. Furthermore, the EndoKED pre-trained vision backbone enables data-efficient and generalisable learning for optical biopsy, achieving expert-level performance in both retrospective and prospective validation.
Rethinking Multiple Instance Learning for Whole Slide Image Classification: A Good Instance Classifier is All You Need
Jul 05, 2023



Abstract:Weakly supervised whole slide image classification is usually formulated as a multiple instance learning (MIL) problem, where each slide is treated as a bag, and the patches cut out of it are treated as instances. Existing methods either train an instance classifier through pseudo-labeling or aggregate instance features into a bag feature through attention mechanisms and then train a bag classifier, where the attention scores can be used for instance-level classification. However, the pseudo instance labels constructed by the former usually contain a lot of noise, and the attention scores constructed by the latter are not accurate enough, both of which affect their performance. In this paper, we propose an instance-level MIL framework based on contrastive learning and prototype learning to effectively accomplish both instance classification and bag classification tasks. To this end, we propose an instance-level weakly supervised contrastive learning algorithm for the first time under the MIL setting to effectively learn instance feature representation. We also propose an accurate pseudo label generation method through prototype learning. We then develop a joint training strategy for weakly supervised contrastive learning, prototype learning, and instance classifier training. Extensive experiments and visualizations on four datasets demonstrate the powerful performance of our method. Codes will be available.
The Rise of AI Language Pathologists: Exploring Two-level Prompt Learning for Few-shot Weakly-supervised Whole Slide Image Classification
May 29, 2023



Abstract:This paper introduces the novel concept of few-shot weakly supervised learning for pathology Whole Slide Image (WSI) classification, denoted as FSWC. A solution is proposed based on prompt learning and the utilization of a large language model, GPT-4. Since a WSI is too large and needs to be divided into patches for processing, WSI classification is commonly approached as a Multiple Instance Learning (MIL) problem. In this context, each WSI is considered a bag, and the obtained patches are treated as instances. The objective of FSWC is to classify both bags and instances with only a limited number of labeled bags. Unlike conventional few-shot learning problems, FSWC poses additional challenges due to its weak bag labels within the MIL framework. Drawing inspiration from the recent achievements of vision-language models (V-L models) in downstream few-shot classification tasks, we propose a two-level prompt learning MIL framework tailored for pathology, incorporating language prior knowledge. Specifically, we leverage CLIP to extract instance features for each patch, and introduce a prompt-guided pooling strategy to aggregate these instance features into a bag feature. Subsequently, we employ a small number of labeled bags to facilitate few-shot prompt learning based on the bag features. Our approach incorporates the utilization of GPT-4 in a question-and-answer mode to obtain language prior knowledge at both the instance and bag levels, which are then integrated into the instance and bag level language prompts. Additionally, a learnable component of the language prompts is trained using the available few-shot labeled data. We conduct extensive experiments on three real WSI datasets encompassing breast cancer, lung cancer, and cervical cancer, demonstrating the notable performance of the proposed method in bag and instance classification. All codes will be made publicly accessible.
Robust Point Cloud Registration Framework Based on Deep Graph Matching(TPAMI Version)
Nov 09, 2022



Abstract:3D point cloud registration is a fundamental problem in computer vision and robotics. Recently, learning-based point cloud registration methods have made great progress. However, these methods are sensitive to outliers, which lead to more incorrect correspondences. In this paper, we propose a novel deep graph matching-based framework for point cloud registration. Specifically, we first transform point clouds into graphs and extract deep features for each point. Then, we develop a module based on deep graph matching to calculate a soft correspondence matrix. By using graph matching, not only the local geometry of each point but also its structure and topology in a larger range are considered in establishing correspondences, so that more correct correspondences are found. We train the network with a loss directly defined on the correspondences, and in the test stage the soft correspondences are transformed into hard one-to-one correspondences so that registration can be performed by a correspondence-based solver. Furthermore, we introduce a transformer-based method to generate edges for graph construction, which further improves the quality of the correspondences. Extensive experiments on object-level and scene-level benchmark datasets show that the proposed method achieves state-of-the-art performance. The code is available at: \href{https://github.com/fukexue/RGM}{https://github.com/fukexue/RGM}.
Bi-directional Weakly Supervised Knowledge Distillation for Whole Slide Image Classification
Oct 10, 2022



Abstract:Computer-aided pathology diagnosis based on the classification of Whole Slide Image (WSI) plays an important role in clinical practice, and it is often formulated as a weakly-supervised Multiple Instance Learning (MIL) problem. Existing methods solve this problem from either a bag classification or an instance classification perspective. In this paper, we propose an end-to-end weakly supervised knowledge distillation framework (WENO) for WSI classification, which integrates a bag classifier and an instance classifier in a knowledge distillation framework to mutually improve the performance of both classifiers. Specifically, an attention-based bag classifier is used as the teacher network, which is trained with weak bag labels, and an instance classifier is used as the student network, which is trained using the normalized attention scores obtained from the teacher network as soft pseudo labels for the instances in positive bags. An instance feature extractor is shared between the teacher and the student to further enhance the knowledge exchange between them. In addition, we propose a hard positive instance mining strategy based on the output of the student network to force the teacher network to keep mining hard positive instances. WENO is a plug-and-play framework that can be easily applied to any existing attention-based bag classification methods. Extensive experiments on five datasets demonstrate the efficiency of WENO. Code is available at https://github.com/miccaiif/WENO.
DGMIL: Distribution Guided Multiple Instance Learning for Whole Slide Image Classification
Jun 17, 2022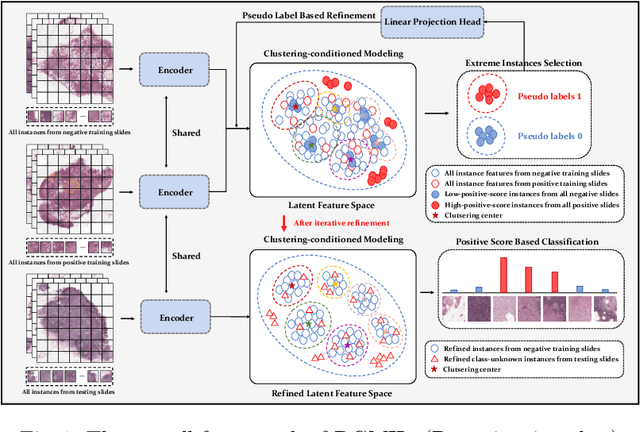
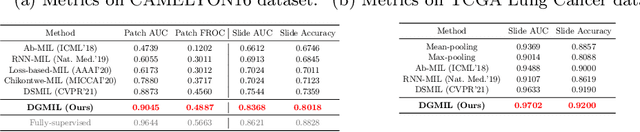

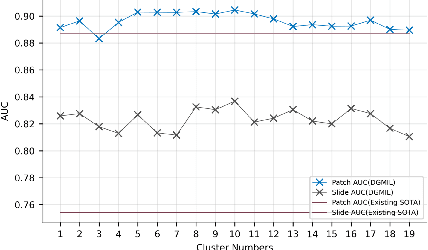
Abstract:Multiple Instance Learning (MIL) is widely used in analyzing histopathological Whole Slide Images (WSIs). However, existing MIL methods do not explicitly model the data distribution, and instead they only learn a bag-level or instance-level decision boundary discriminatively by training a classifier. In this paper, we propose DGMIL: a feature distribution guided deep MIL framework for WSI classification and positive patch localization. Instead of designing complex discriminative network architectures, we reveal that the inherent feature distribution of histopathological image data can serve as a very effective guide for instance classification. We propose a cluster-conditioned feature distribution modeling method and a pseudo label-based iterative feature space refinement strategy so that in the final feature space the positive and negative instances can be easily separated. Experiments on the CAMELYON16 dataset and the TCGA Lung Cancer dataset show that our method achieves new SOTA for both global classification and positive patch localization tasks.
Multi-View Partial Point Cloud Challenge 2021 on Completion and Registration: Methods and Results
Dec 22, 2021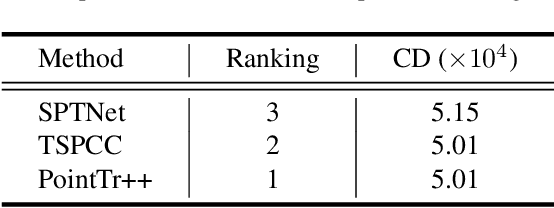
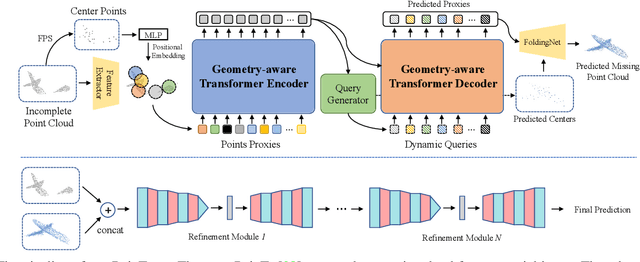
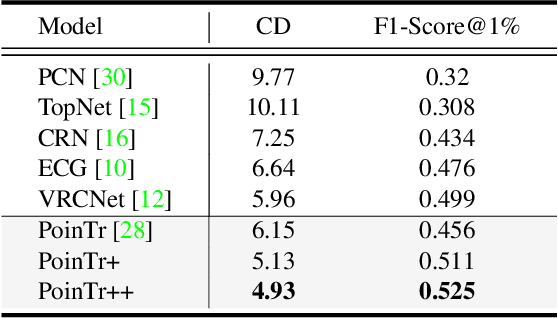
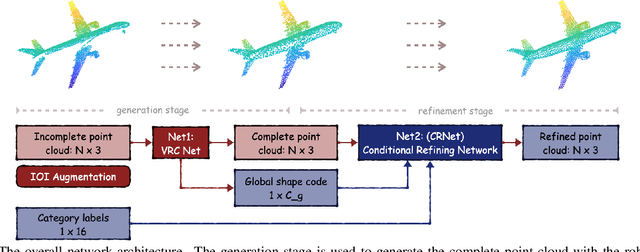
Abstract:As real-scanned point clouds are mostly partial due to occlusions and viewpoints, reconstructing complete 3D shapes based on incomplete observations becomes a fundamental problem for computer vision. With a single incomplete point cloud, it becomes the partial point cloud completion problem. Given multiple different observations, 3D reconstruction can be addressed by performing partial-to-partial point cloud registration. Recently, a large-scale Multi-View Partial (MVP) point cloud dataset has been released, which consists of over 100,000 high-quality virtual-scanned partial point clouds. Based on the MVP dataset, this paper reports methods and results in the Multi-View Partial Point Cloud Challenge 2021 on Completion and Registration. In total, 128 participants registered for the competition, and 31 teams made valid submissions. The top-ranked solutions will be analyzed, and then we will discuss future research directions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge