Xinrong Chen
Residual Decoding: Mitigating Hallucinations in Large Vision-Language Models via History-Aware Residual Guidance
Feb 01, 2026Abstract:Large Vision-Language Models (LVLMs) can reason effectively from image-text inputs and perform well in various multimodal tasks. Despite this success, they are affected by language priors and often produce hallucinations. Hallucinations denote generated content that is grammatically and syntactically coherent, yet bears no match or direct relevance to actual visual input. To address this problem, we propose Residual Decoding (ResDec). It is a novel training-free method that uses historical information to aid decoding. The method relies on the internal implicit reasoning mechanism and token logits evolution mechanism of LVLMs to correct biases. Extensive experiments demonstrate that ResDec effectively suppresses hallucinations induced by language priors, significantly improves visual grounding, and reduces object hallucinations. In addition to mitigating hallucinations, ResDec also performs exceptionally well on comprehensive LVLM benchmarks, highlighting its broad applicability.
KineST: A Kinematics-guided Spatiotemporal State Space Model for Human Motion Tracking from Sparse Signals
Dec 18, 2025Abstract:Full-body motion tracking plays an essential role in AR/VR applications, bridging physical and virtual interactions. However, it is challenging to reconstruct realistic and diverse full-body poses based on sparse signals obtained by head-mounted displays, which are the main devices in AR/VR scenarios. Existing methods for pose reconstruction often incur high computational costs or rely on separately modeling spatial and temporal dependencies, making it difficult to balance accuracy, temporal coherence, and efficiency. To address this problem, we propose KineST, a novel kinematics-guided state space model, which effectively extracts spatiotemporal dependencies while integrating local and global pose perception. The innovation comes from two core ideas. Firstly, in order to better capture intricate joint relationships, the scanning strategy within the State Space Duality framework is reformulated into kinematics-guided bidirectional scanning, which embeds kinematic priors. Secondly, a mixed spatiotemporal representation learning approach is employed to tightly couple spatial and temporal contexts, balancing accuracy and smoothness. Additionally, a geometric angular velocity loss is introduced to impose physically meaningful constraints on rotational variations for further improving motion stability. Extensive experiments demonstrate that KineST has superior performance in both accuracy and temporal consistency within a lightweight framework. Project page: https://kaka-1314.github.io/KineST/
GuiLoMo: Allocating Expert Number and Rank for LoRA-MoE via Bilevel Optimization with GuidedSelection Vectors
Jun 17, 2025Abstract:Parameter-efficient fine-tuning (PEFT) methods, particularly Low-Rank Adaptation (LoRA), offer an efficient way to adapt large language models with reduced computational costs. However, their performance is limited by the small number of trainable parameters. Recent work combines LoRA with the Mixture-of-Experts (MoE), i.e., LoRA-MoE, to enhance capacity, but two limitations remain in hindering the full exploitation of its potential: 1) the influence of downstream tasks when assigning expert numbers, and 2) the uniform rank assignment across all LoRA experts, which restricts representational diversity. To mitigate these gaps, we propose GuiLoMo, a fine-grained layer-wise expert numbers and ranks allocation strategy with GuidedSelection Vectors (GSVs). GSVs are learned via a prior bilevel optimization process to capture both model- and task-specific needs, and are then used to allocate optimal expert numbers and ranks. Experiments on three backbone models across diverse benchmarks show that GuiLoMo consistently achieves superior or comparable performance to all baselines. Further analysis offers key insights into how expert numbers and ranks vary across layers and tasks, highlighting the benefits of adaptive expert configuration. Our code is available at https://github.com/Liar406/Gui-LoMo.git.
Qwen Look Again: Guiding Vision-Language Reasoning Models to Re-attention Visual Information
May 29, 2025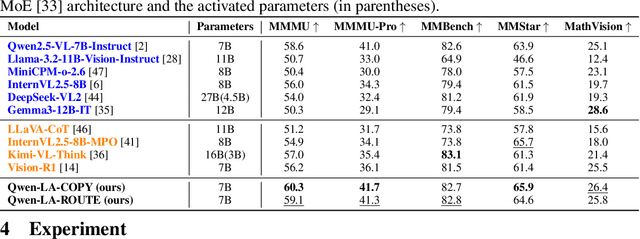
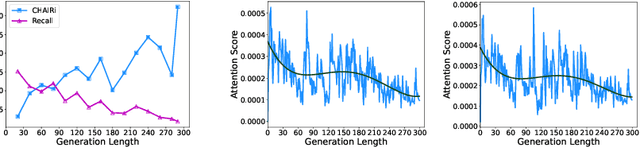
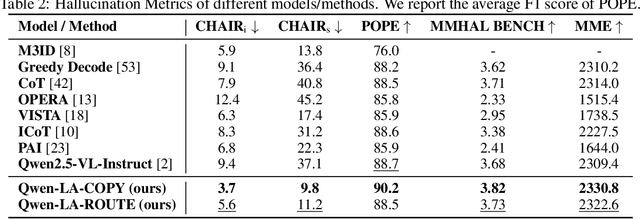
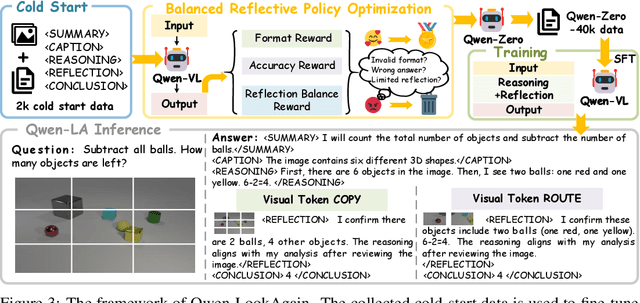
Abstract:Inference time scaling drives extended reasoning to enhance the performance of Vision-Language Models (VLMs), thus forming powerful Vision-Language Reasoning Models (VLRMs). However, long reasoning dilutes visual tokens, causing visual information to receive less attention and may trigger hallucinations. Although introducing text-only reflection processes shows promise in language models, we demonstrate that it is insufficient to suppress hallucinations in VLMs. To address this issue, we introduce Qwen-LookAgain (Qwen-LA), a novel VLRM designed to mitigate hallucinations by incorporating a vision-text reflection process that guides the model to re-attention visual information during reasoning. We first propose a reinforcement learning method Balanced Reflective Policy Optimization (BRPO), which guides the model to decide when to generate vision-text reflection on its own and balance the number and length of reflections. Then, we formally prove that VLRMs lose attention to visual tokens as reasoning progresses, and demonstrate that supplementing visual information during reflection enhances visual attention. Therefore, during training and inference, Visual Token COPY and Visual Token ROUTE are introduced to force the model to re-attention visual information at the visual level, addressing the limitations of text-only reflection. Experiments on multiple visual QA datasets and hallucination metrics indicate that Qwen-LA achieves leading accuracy performance while reducing hallucinations. Our code is available at: https://github.com/Liar406/Look_Again.
SSD-Poser: Avatar Pose Estimation with State Space Duality from Sparse Observations
Apr 25, 2025Abstract:The growing applications of AR/VR increase the demand for real-time full-body pose estimation from Head-Mounted Displays (HMDs). Although HMDs provide joint signals from the head and hands, reconstructing a full-body pose remains challenging due to the unconstrained lower body. Recent advancements often rely on conventional neural networks and generative models to improve performance in this task, such as Transformers and diffusion models. However, these approaches struggle to strike a balance between achieving precise pose reconstruction and maintaining fast inference speed. To overcome these challenges, a lightweight and efficient model, SSD-Poser, is designed for robust full-body motion estimation from sparse observations. SSD-Poser incorporates a well-designed hybrid encoder, State Space Attention Encoders, to adapt the state space duality to complex motion poses and enable real-time realistic pose reconstruction. Moreover, a Frequency-Aware Decoder is introduced to mitigate jitter caused by variable-frequency motion signals, remarkably enhancing the motion smoothness. Comprehensive experiments on the AMASS dataset demonstrate that SSD-Poser achieves exceptional accuracy and computational efficiency, showing outstanding inference efficiency compared to state-of-the-art methods.
REMOTE: Real-time Ego-motion Tracking for Various Endoscopes via Multimodal Visual Feature Learning
Jan 30, 2025Abstract:Real-time ego-motion tracking for endoscope is a significant task for efficient navigation and robotic automation of endoscopy. In this paper, a novel framework is proposed to perform real-time ego-motion tracking for endoscope. Firstly, a multi-modal visual feature learning network is proposed to perform relative pose prediction, in which the motion feature from the optical flow, the scene features and the joint feature from two adjacent observations are all extracted for prediction. Due to more correlation information in the channel dimension of the concatenated image, a novel feature extractor is designed based on an attention mechanism to integrate multi-dimensional information from the concatenation of two continuous frames. To extract more complete feature representation from the fused features, a novel pose decoder is proposed to predict the pose transformation from the concatenated feature map at the end of the framework. At last, the absolute pose of endoscope is calculated based on relative poses. The experiment is conducted on three datasets of various endoscopic scenes and the results demonstrate that the proposed method outperforms state-of-the-art methods. Besides, the inference speed of the proposed method is over 30 frames per second, which meets the real-time requirement. The project page is here: \href{https://remote-bmxs.netlify.app}{remote-bmxs.netlify.app}
Towards Full-parameter and Parameter-efficient Self-learning For Endoscopic Camera Depth Estimation
Oct 01, 2024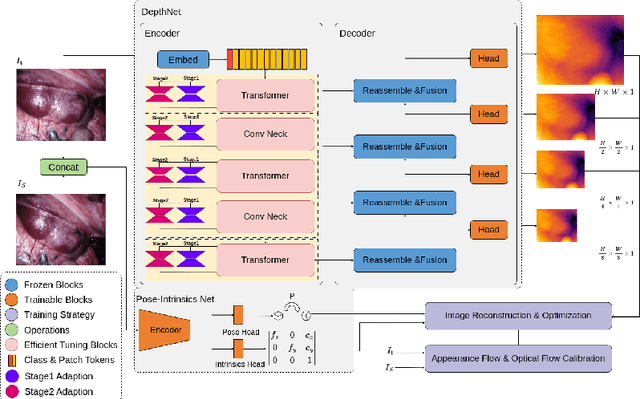
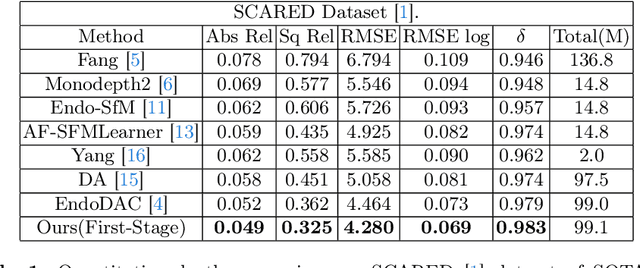
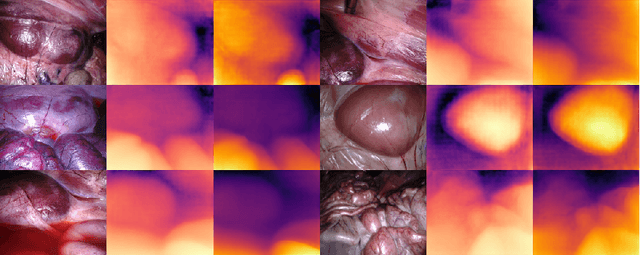
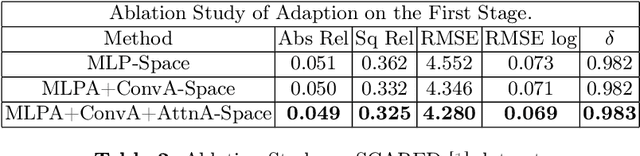
Abstract:Adaptation methods are developed to adapt depth foundation models to endoscopic depth estimation recently. However, such approaches typically under-perform training since they limit the parameter search to a low-rank subspace and alter the training dynamics. Therefore, we propose a full-parameter and parameter-efficient learning framework for endoscopic depth estimation. At the first stage, the subspace of attention, convolution and multi-layer perception are adapted simultaneously within different sub-spaces. At the second stage, a memory-efficient optimization is proposed for subspace composition and the performance is further improved in the united sub-space. Initial experiments on the SCARED dataset demonstrate that results at the first stage improves the performance from 10.2% to 4.1% for Sq Rel, Abs Rel, RMSE and RMSE log in the comparison with the state-of-the-art models.
Rethinking Multiple Instance Learning: Developing an Instance-Level Classifier via Weakly-Supervised Self-Training
Aug 09, 2024Abstract:Multiple instance learning (MIL) problem is currently solved from either bag-classification or instance-classification perspective, both of which ignore important information contained in some instances and result in limited performance. For example, existing methods often face difficulty in learning hard positive instances. In this paper, we formulate MIL as a semi-supervised instance classification problem, so that all the labeled and unlabeled instances can be fully utilized to train a better classifier. The difficulty in this formulation is that all the labeled instances are negative in MIL, and traditional self-training techniques used in semi-supervised learning tend to degenerate in generating pseudo labels for the unlabeled instances in this scenario. To resolve this problem, we propose a weakly-supervised self-training method, in which we utilize the positive bag labels to construct a global constraint and a local constraint on the pseudo labels to prevent them from degenerating and force the classifier to learn hard positive instances. It is worth noting that easy positive instances are instances are far from the decision boundary in the classification process, while hard positive instances are those close to the decision boundary. Through iterative optimization, the pseudo labels can gradually approach the true labels. Extensive experiments on two MNIST synthetic datasets, five traditional MIL benchmark datasets and two histopathology whole slide image datasets show that our method achieved new SOTA performance on all of them. The code will be publicly available.
The Extreme Cardiac MRI Analysis Challenge under Respiratory Motion (CMRxMotion)
Oct 12, 2022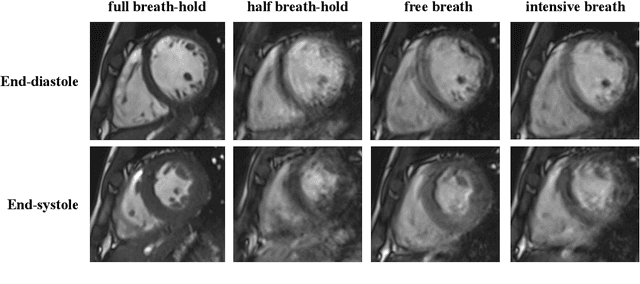
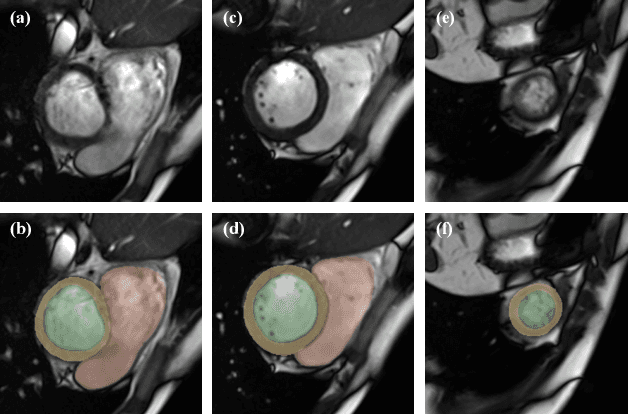

Abstract:The quality of cardiac magnetic resonance (CMR) imaging is susceptible to respiratory motion artifacts. The model robustness of automated segmentation techniques in face of real-world respiratory motion artifacts is unclear. This manuscript describes the design of extreme cardiac MRI analysis challenge under respiratory motion (CMRxMotion Challenge). The challenge aims to establish a public benchmark dataset to assess the effects of respiratory motion on image quality and examine the robustness of segmentation models. The challenge recruited 40 healthy volunteers to perform different breath-hold behaviors during one imaging visit, obtaining paired cine imaging with artifacts. Radiologists assessed the image quality and annotated the level of respiratory motion artifacts. For those images with diagnostic quality, radiologists further segmented the left ventricle, left ventricle myocardium and right ventricle. The images of training set (20 volunteers) along with the annotations are released to the challenge participants, to develop an automated image quality assessment model (Task 1) and an automated segmentation model (Task 2). The images of validation set (5 volunteers) are released to the challenge participants but the annotations are withheld for online evaluation of submitted predictions. Both the images and annotations of the test set (15 volunteers) were withheld and only used for offline evaluation of submitted containerized dockers. The image quality assessment task is quantitatively evaluated by the Cohen's kappa statistics and the segmentation task is evaluated by the Dice scores and Hausdorff distances.
A Benchmark for Weakly Semi-Supervised Abnormality Localization in Chest X-Rays
Sep 05, 2022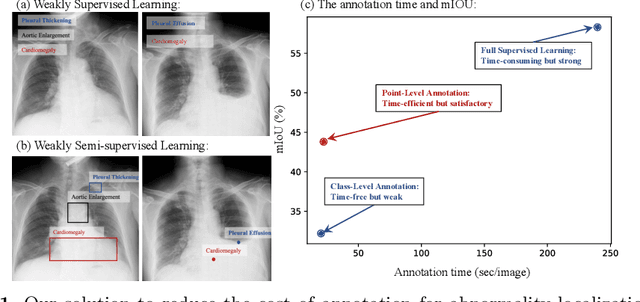
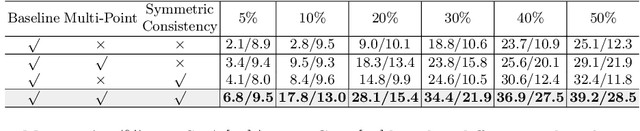
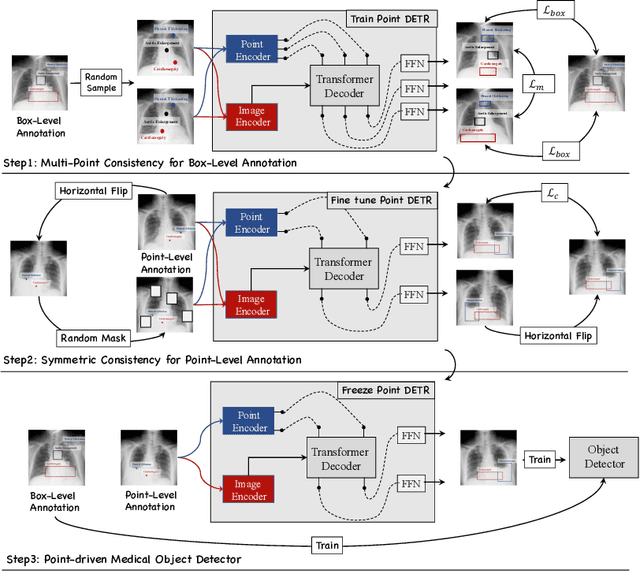
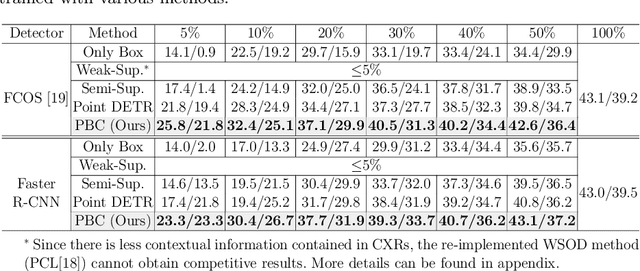
Abstract:Accurate abnormality localization in chest X-rays (CXR) can benefit the clinical diagnosis of various thoracic diseases. However, the lesion-level annotation can only be performed by experienced radiologists, and it is tedious and time-consuming, thus difficult to acquire. Such a situation results in a difficulty to develop a fully-supervised abnormality localization system for CXR. In this regard, we propose to train the CXR abnormality localization framework via a weakly semi-supervised strategy, termed Point Beyond Class (PBC), which utilizes a small number of fully annotated CXRs with lesion-level bounding boxes and extensive weakly annotated samples by points. Such a point annotation setting can provide weakly instance-level information for abnormality localization with a marginal annotation cost. Particularly, the core idea behind our PBC is to learn a robust and accurate mapping from the point annotations to the bounding boxes against the variance of annotated points. To achieve that, a regularization term, namely multi-point consistency, is proposed, which drives the model to generate the consistent bounding box from different point annotations inside the same abnormality. Furthermore, a self-supervision, termed symmetric consistency, is also proposed to deeply exploit the useful information from the weakly annotated data for abnormality localization. Experimental results on RSNA and VinDr-CXR datasets justify the effectiveness of the proposed method. When less than 20% box-level labels are used for training, an improvement of ~5 in mAP can be achieved by our PBC, compared to the current state-of-the-art method (i.e., Point DETR). Code is available at https://github.com/HaozheLiu-ST/Point-Beyond-Class.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge