Nanjun He
Cross-Modal Vertical Federated Learning for MRI Reconstruction
Jun 05, 2023Abstract:Federated learning enables multiple hospitals to cooperatively learn a shared model without privacy disclosure. Existing methods often take a common assumption that the data from different hospitals have the same modalities. However, such a setting is difficult to fully satisfy in practical applications, since the imaging guidelines may be different between hospitals, which makes the number of individuals with the same set of modalities limited. To this end, we formulate this practical-yet-challenging cross-modal vertical federated learning task, in which shape data from multiple hospitals have different modalities with a small amount of multi-modality data collected from the same individuals. To tackle such a situation, we develop a novel framework, namely Federated Consistent Regularization constrained Feature Disentanglement (Fed-CRFD), for boosting MRI reconstruction by effectively exploring the overlapping samples (individuals with multi-modalities) and solving the domain shift problem caused by different modalities. Particularly, our Fed-CRFD involves an intra-client feature disentangle scheme to decouple data into modality-invariant and modality-specific features, where the modality-invariant features are leveraged to mitigate the domain shift problem. In addition, a cross-client latent representation consistency constraint is proposed specifically for the overlapping samples to further align the modality-invariant features extracted from different modalities. Hence, our method can fully exploit the multi-source data from hospitals while alleviating the domain shift problem. Extensive experiments on two typical MRI datasets demonstrate that our network clearly outperforms state-of-the-art MRI reconstruction methods. The source code will be publicly released upon the publication of this work.
Improving GAN Training via Feature Space Shrinkage
Mar 02, 2023Abstract:Due to the outstanding capability for data generation, Generative Adversarial Networks (GANs) have attracted considerable attention in unsupervised learning. However, training GANs is difficult, since the training distribution is dynamic for the discriminator, leading to unstable image representation. In this paper, we address the problem of training GANs from a novel perspective, \emph{i.e.,} robust image classification. Motivated by studies on robust image representation, we propose a simple yet effective module, namely AdaptiveMix, for GANs, which shrinks the regions of training data in the image representation space of the discriminator. Considering it is intractable to directly bound feature space, we propose to construct hard samples and narrow down the feature distance between hard and easy samples. The hard samples are constructed by mixing a pair of training images. We evaluate the effectiveness of our AdaptiveMix with widely-used and state-of-the-art GAN architectures. The evaluation results demonstrate that our AdaptiveMix can facilitate the training of GANs and effectively improve the image quality of generated samples. We also show that our AdaptiveMix can be further applied to image classification and Out-Of-Distribution (OOD) detection tasks, by equipping it with state-of-the-art methods. Extensive experiments on seven publicly available datasets show that our method effectively boosts the performance of baselines. The code is publicly available at https://github.com/WentianZhang-ML/AdaptiveMix.
A New Perspective to Boost Vision Transformer for Medical Image Classification
Jan 03, 2023Abstract:Transformer has achieved impressive successes for various computer vision tasks. However, most of existing studies require to pretrain the Transformer backbone on a large-scale labeled dataset (e.g., ImageNet) for achieving satisfactory performance, which is usually unavailable for medical images. Additionally, due to the gap between medical and natural images, the improvement generated by the ImageNet pretrained weights significantly degrades while transferring the weights to medical image processing tasks. In this paper, we propose Bootstrap Own Latent of Transformer (BOLT), a self-supervised learning approach specifically for medical image classification with the Transformer backbone. Our BOLT consists of two networks, namely online and target branches, for self-supervised representation learning. Concretely, the online network is trained to predict the target network representation of the same patch embedding tokens with a different perturbation. To maximally excavate the impact of Transformer from limited medical data, we propose an auxiliary difficulty ranking task. The Transformer is enforced to identify which branch (i.e., online/target) is processing the more difficult perturbed tokens. Overall, the Transformer endeavours itself to distill the transformation-invariant features from the perturbed tokens to simultaneously achieve difficulty measurement and maintain the consistency of self-supervised representations. The proposed BOLT is evaluated on three medical image processing tasks, i.e., skin lesion classification, knee fatigue fracture grading and diabetic retinopathy grading. The experimental results validate the superiority of our BOLT for medical image classification, compared to ImageNet pretrained weights and state-of-the-art self-supervised learning approaches.
A Benchmark for Weakly Semi-Supervised Abnormality Localization in Chest X-Rays
Sep 05, 2022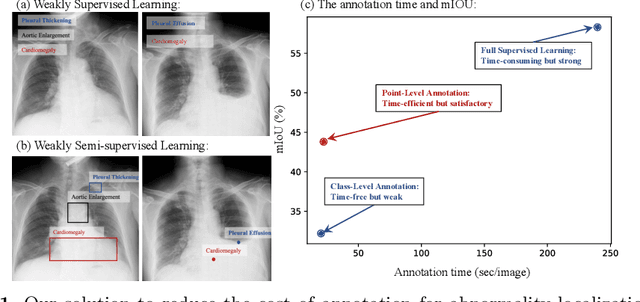
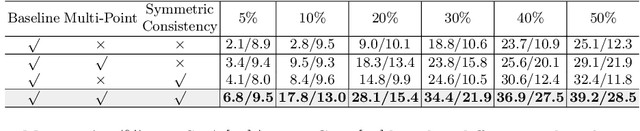
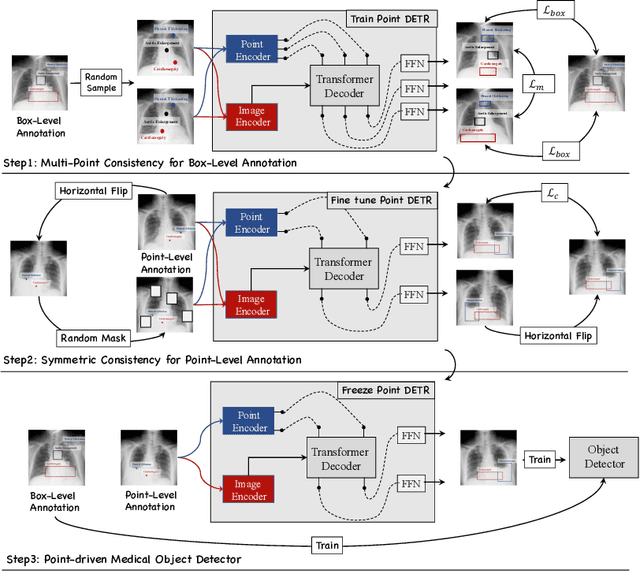
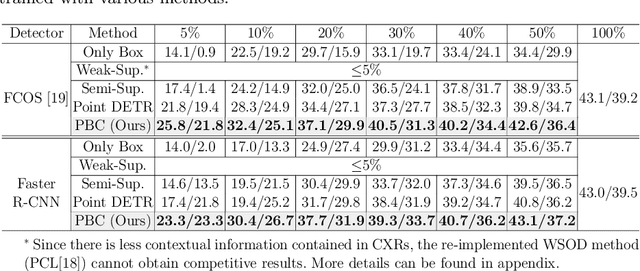
Abstract:Accurate abnormality localization in chest X-rays (CXR) can benefit the clinical diagnosis of various thoracic diseases. However, the lesion-level annotation can only be performed by experienced radiologists, and it is tedious and time-consuming, thus difficult to acquire. Such a situation results in a difficulty to develop a fully-supervised abnormality localization system for CXR. In this regard, we propose to train the CXR abnormality localization framework via a weakly semi-supervised strategy, termed Point Beyond Class (PBC), which utilizes a small number of fully annotated CXRs with lesion-level bounding boxes and extensive weakly annotated samples by points. Such a point annotation setting can provide weakly instance-level information for abnormality localization with a marginal annotation cost. Particularly, the core idea behind our PBC is to learn a robust and accurate mapping from the point annotations to the bounding boxes against the variance of annotated points. To achieve that, a regularization term, namely multi-point consistency, is proposed, which drives the model to generate the consistent bounding box from different point annotations inside the same abnormality. Furthermore, a self-supervision, termed symmetric consistency, is also proposed to deeply exploit the useful information from the weakly annotated data for abnormality localization. Experimental results on RSNA and VinDr-CXR datasets justify the effectiveness of the proposed method. When less than 20% box-level labels are used for training, an improvement of ~5 in mAP can be achieved by our PBC, compared to the current state-of-the-art method (i.e., Point DETR). Code is available at https://github.com/HaozheLiu-ST/Point-Beyond-Class.
mmFormer: Multimodal Medical Transformer for Incomplete Multimodal Learning of Brain Tumor Segmentation
Jun 06, 2022



Abstract:Accurate brain tumor segmentation from Magnetic Resonance Imaging (MRI) is desirable to joint learning of multimodal images. However, in clinical practice, it is not always possible to acquire a complete set of MRIs, and the problem of missing modalities causes severe performance degradation in existing multimodal segmentation methods. In this work, we present the first attempt to exploit the Transformer for multimodal brain tumor segmentation that is robust to any combinatorial subset of available modalities. Concretely, we propose a novel multimodal Medical Transformer (mmFormer) for incomplete multimodal learning with three main components: the hybrid modality-specific encoders that bridge a convolutional encoder and an intra-modal Transformer for both local and global context modeling within each modality; an inter-modal Transformer to build and align the long-range correlations across modalities for modality-invariant features with global semantics corresponding to tumor region; a decoder that performs a progressive up-sampling and fusion with the modality-invariant features to generate robust segmentation. Besides, auxiliary regularizers are introduced in both encoder and decoder to further enhance the model's robustness to incomplete modalities. We conduct extensive experiments on the public BraTS $2018$ dataset for brain tumor segmentation. The results demonstrate that the proposed mmFormer outperforms the state-of-the-art methods for incomplete multimodal brain tumor segmentation on almost all subsets of incomplete modalities, especially by an average 19.07% improvement of Dice on tumor segmentation with only one available modality. The code is available at https://github.com/YaoZhang93/mmFormer.
Robust Representation via Dynamic Feature Aggregation
May 16, 2022



Abstract:Deep convolutional neural network (CNN) based models are vulnerable to the adversarial attacks. One of the possible reasons is that the embedding space of CNN based model is sparse, resulting in a large space for the generation of adversarial samples. In this study, we propose a method, denoted as Dynamic Feature Aggregation, to compress the embedding space with a novel regularization. Particularly, the convex combination between two samples are regarded as the pivot for aggregation. In the embedding space, the selected samples are guided to be similar to the representation of the pivot. On the other side, to mitigate the trivial solution of such regularization, the last fully-connected layer of the model is replaced by an orthogonal classifier, in which the embedding codes for different classes are processed orthogonally and separately. With the regularization and orthogonal classifier, a more compact embedding space can be obtained, which accordingly improves the model robustness against adversarial attacks. An averaging accuracy of 56.91% is achieved by our method on CIFAR-10 against various attack methods, which significantly surpasses a solid baseline (Mixup) by a margin of 37.31%. More surprisingly, empirical results show that, the proposed method can also achieve the state-of-the-art performance for out-of-distribution (OOD) detection, due to the learned compact feature space. An F1 score of 0.937 is achieved by the proposed method, when adopting CIFAR-10 as in-distribution (ID) dataset and LSUN as OOD dataset. Code is available at https://github.com/HaozheLiu-ST/DynamicFeatureAggregation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge