Kui Huang
Qwen Look Again: Guiding Vision-Language Reasoning Models to Re-attention Visual Information
May 29, 2025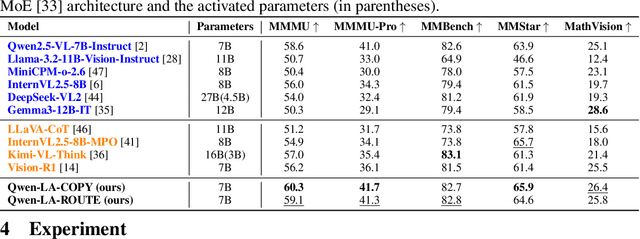
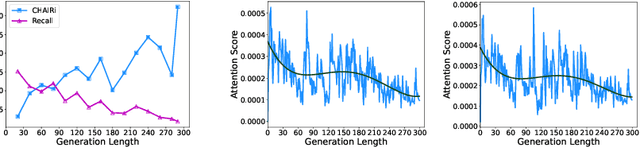
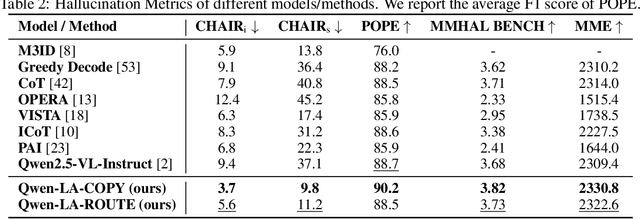
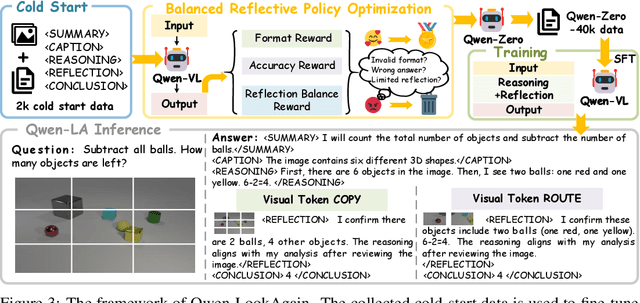
Abstract:Inference time scaling drives extended reasoning to enhance the performance of Vision-Language Models (VLMs), thus forming powerful Vision-Language Reasoning Models (VLRMs). However, long reasoning dilutes visual tokens, causing visual information to receive less attention and may trigger hallucinations. Although introducing text-only reflection processes shows promise in language models, we demonstrate that it is insufficient to suppress hallucinations in VLMs. To address this issue, we introduce Qwen-LookAgain (Qwen-LA), a novel VLRM designed to mitigate hallucinations by incorporating a vision-text reflection process that guides the model to re-attention visual information during reasoning. We first propose a reinforcement learning method Balanced Reflective Policy Optimization (BRPO), which guides the model to decide when to generate vision-text reflection on its own and balance the number and length of reflections. Then, we formally prove that VLRMs lose attention to visual tokens as reasoning progresses, and demonstrate that supplementing visual information during reflection enhances visual attention. Therefore, during training and inference, Visual Token COPY and Visual Token ROUTE are introduced to force the model to re-attention visual information at the visual level, addressing the limitations of text-only reflection. Experiments on multiple visual QA datasets and hallucination metrics indicate that Qwen-LA achieves leading accuracy performance while reducing hallucinations. Our code is available at: https://github.com/Liar406/Look_Again.
PP-DocBee: Improving Multimodal Document Understanding Through a Bag of Tricks
Mar 06, 2025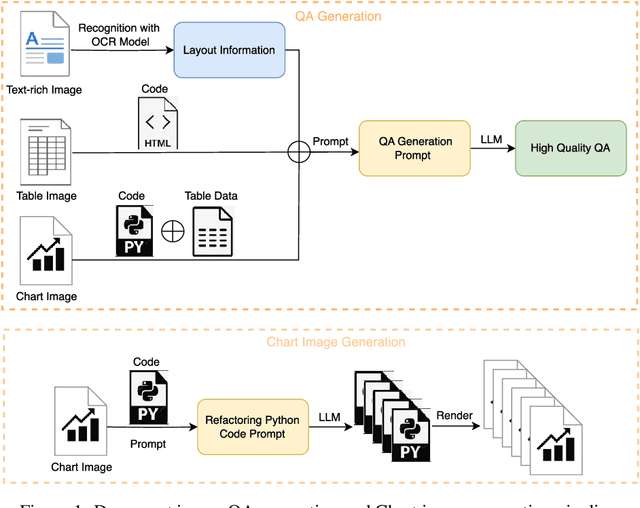

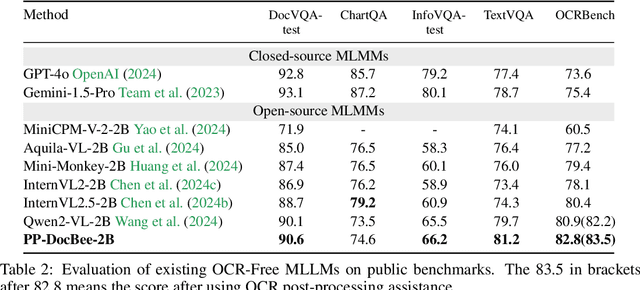
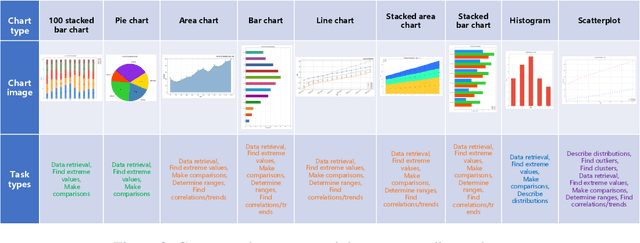
Abstract:With the rapid advancement of digitalization, various document images are being applied more extensively in production and daily life, and there is an increasingly urgent need for fast and accurate parsing of the content in document images. Therefore, this report presents PP-DocBee, a novel multimodal large language model designed for end-to-end document image understanding. First, we develop a data synthesis strategy tailored to document scenarios in which we build a diverse dataset to improve the model generalization. Then, we apply a few training techniques, including dynamic proportional sampling, data preprocessing, and OCR postprocessing strategies. Extensive evaluations demonstrate the superior performance of PP-DocBee, achieving state-of-the-art results on English document understanding benchmarks and even outperforming existing open source and commercial models in Chinese document understanding. The source code and pre-trained models are publicly available at \href{https://github.com/PaddlePaddle/PaddleMIX}{https://github.com/PaddlePaddle/PaddleMIX}.
Unsupervised Waste Classification By Dual-Encoder Contrastive Learning and Multi-Clustering Voting (DECMCV)
Mar 04, 2025



Abstract:Waste classification is crucial for improving processing efficiency and reducing environmental pollution. Supervised deep learning methods are commonly used for automated waste classification, but they rely heavily on large labeled datasets, which are costly and inefficient to obtain. Real-world waste data often exhibit category and style biases, such as variations in camera angles, lighting conditions, and types of waste, which can impact the model's performance and generalization ability. Therefore, constructing a bias-free dataset is essential. Manual labeling is not only costly but also inefficient. While self-supervised learning helps address data scarcity, it still depends on some labeled data and generally results in lower accuracy compared to supervised methods. Unsupervised methods show potential in certain cases but typically do not perform as well as supervised models, highlighting the need for an efficient and cost-effective unsupervised approach. This study presents a novel unsupervised method, Dual-Encoder Contrastive Learning with Multi-Clustering Voting (DECMCV). The approach involves using a pre-trained ConvNeXt model for image encoding, leveraging VisionTransformer to generate positive samples, and applying a multi-clustering voting mechanism to address data labeling and domain shift issues. Experimental results demonstrate that DECMCV achieves classification accuracies of 93.78% and 98.29% on the TrashNet and Huawei Cloud datasets, respectively, outperforming or matching supervised models. On a real-world dataset of 4,169 waste images, only 50 labeled samples were needed to accurately label thousands, improving classification accuracy by 29.85% compared to supervised models. This method effectively addresses style differences, enhances model generalization, and contributes to the advancement of automated waste classification.
Digital Modeling of Massage Techniques and Reproduction by Robotic Arms
Dec 08, 2024Abstract:This paper explores the digital modeling and robotic reproduction of traditional Chinese medicine (TCM) massage techniques. We adopt an adaptive admittance control algorithm to optimize force and position control, ensuring safety and comfort. The paper analyzes key TCM techniques from kinematic and dynamic perspectives, and designs robotic systems to reproduce these massage techniques. The results demonstrate that the robot successfully mimics the characteristics of TCM massage, providing a foundation for integrating traditional therapy with modern robotics and expanding assistive therapy applications.
RT-DETRv2: Improved Baseline with Bag-of-Freebies for Real-Time Detection Transformer
Jul 24, 2024Abstract:In this report, we present RT-DETRv2, an improved Real-Time DEtection TRansformer (RT-DETR). RT-DETRv2 builds upon the previous state-of-the-art real-time detector, RT-DETR, and opens up a set of bag-of-freebies for flexibility and practicality, as well as optimizing the training strategy to achieve enhanced performance. To improve the flexibility, we suggest setting a distinct number of sampling points for features at different scales in the deformable attention to achieve selective multi-scale feature extraction by the decoder. To enhance practicality, we propose an optional discrete sampling operator to replace the grid_sample operator that is specific to RT-DETR compared to YOLOs. This removes the deployment constraints typically associated with DETRs. For the training strategy, we propose dynamic data augmentation and scale-adaptive hyperparameters customization to improve performance without loss of speed. Source code and pre-trained models will be available at https://github.com/lyuwenyu/RT-DETR.
Empowering AI drug discovery with explicit and implicit knowledge
Apr 17, 2023Abstract:Motivation: Recently, research on independently utilizing either explicit knowledge from knowledge graphs or implicit knowledge from biomedical literature for AI drug discovery has been growing rapidly. These approaches have greatly improved the prediction accuracy of AI models on multiple downstream tasks. However, integrating explicit and implicit knowledge independently hinders their understanding of molecules. Results: We propose DeepEIK, a unified deep learning framework that incorporates both explicit and implicit knowledge for AI drug discovery. We adopt feature fusion to process the multi-modal inputs, and leverage the attention mechanism to denoise the text information. Experiments show that DeepEIK significantly outperforms state-of-the-art methods on crucial tasks in AI drug discovery including drug-target interaction prediction, drug property prediction and protein-protein interaction prediction. Further studies show that benefiting from explicit and implicit knowledge, our framework achieves a deeper understanding of molecules and shows promising potential in facilitating drug discovery applications.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge