Massimo Hong
MutaPLM: Protein Language Modeling for Mutation Explanation and Engineering
Oct 30, 2024



Abstract:Studying protein mutations within amino acid sequences holds tremendous significance in life sciences. Protein language models (PLMs) have demonstrated strong capabilities in broad biological applications. However, due to architectural design and lack of supervision, PLMs model mutations implicitly with evolutionary plausibility, which is not satisfactory to serve as explainable and engineerable tools in real-world studies. To address these issues, we present MutaPLM, a unified framework for interpreting and navigating protein mutations with protein language models. MutaPLM introduces a protein delta network that captures explicit protein mutation representations within a unified feature space, and a transfer learning pipeline with a chain-of-thought (CoT) strategy to harvest protein mutation knowledge from biomedical texts. We also construct MutaDescribe, the first large-scale protein mutation dataset with rich textual annotations, which provides cross-modal supervision signals. Through comprehensive experiments, we demonstrate that MutaPLM excels at providing human-understandable explanations for mutational effects and prioritizing novel mutations with desirable properties. Our code, model, and data are open-sourced at https://github.com/PharMolix/MutaPLM.
Learning Multi-view Molecular Representations with Structured and Unstructured Knowledge
Jun 14, 2024



Abstract:Capturing molecular knowledge with representation learning approaches holds significant potential in vast scientific fields such as chemistry and life science. An effective and generalizable molecular representation is expected to capture the consensus and complementary molecular expertise from diverse views and perspectives. However, existing works fall short in learning multi-view molecular representations, due to challenges in explicitly incorporating view information and handling molecular knowledge from heterogeneous sources. To address these issues, we present MV-Mol, a molecular representation learning model that harvests multi-view molecular expertise from chemical structures, unstructured knowledge from biomedical texts, and structured knowledge from knowledge graphs. We utilize text prompts to model view information and design a fusion architecture to extract view-based molecular representations. We develop a two-stage pre-training procedure, exploiting heterogeneous data of varying quality and quantity. Through extensive experiments, we show that MV-Mol provides improved representations that substantially benefit molecular property prediction. Additionally, MV-Mol exhibits state-of-the-art performance in multi-modal comprehension of molecular structures and texts. Code and data are available at https://github.com/PharMolix/OpenBioMed.
Empowering AI drug discovery with explicit and implicit knowledge
Apr 17, 2023Abstract:Motivation: Recently, research on independently utilizing either explicit knowledge from knowledge graphs or implicit knowledge from biomedical literature for AI drug discovery has been growing rapidly. These approaches have greatly improved the prediction accuracy of AI models on multiple downstream tasks. However, integrating explicit and implicit knowledge independently hinders their understanding of molecules. Results: We propose DeepEIK, a unified deep learning framework that incorporates both explicit and implicit knowledge for AI drug discovery. We adopt feature fusion to process the multi-modal inputs, and leverage the attention mechanism to denoise the text information. Experiments show that DeepEIK significantly outperforms state-of-the-art methods on crucial tasks in AI drug discovery including drug-target interaction prediction, drug property prediction and protein-protein interaction prediction. Further studies show that benefiting from explicit and implicit knowledge, our framework achieves a deeper understanding of molecules and shows promising potential in facilitating drug discovery applications.
Attacks and Faults Injection in Self-Driving Agents on the Carla Simulator -- Experience Report
Feb 25, 2022
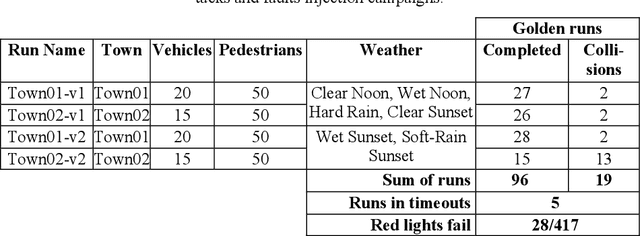
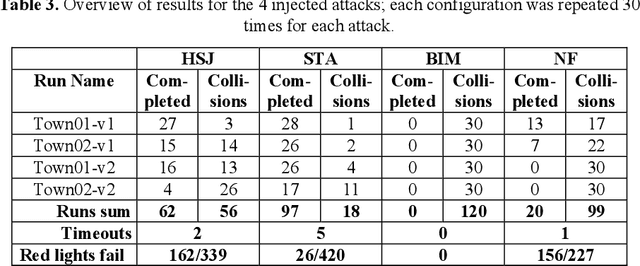
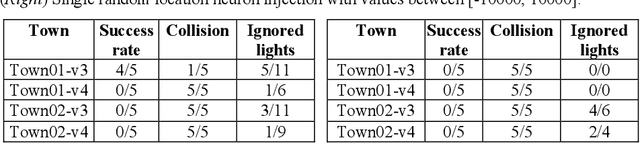
Abstract:Machine Learning applications are acknowledged at the foundation of autonomous driving, because they are the enabling technology for most driving tasks. However, the inclusion of trained agents in automotive systems exposes the vehicle to novel attacks and faults, that can result in safety threats to the driv-ing tasks. In this paper we report our experimental campaign on the injection of adversarial attacks and software faults in a self-driving agent running in a driving simulator. We show that adversarial attacks and faults injected in the trained agent can lead to erroneous decisions and severely jeopardize safety. The paper shows a feasible and easily-reproducible approach based on open source simula-tor and tools, and the results clearly motivate the need of both protective measures and extensive testing campaigns.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge