Xing Yi Liu
Spontaneous Informal Speech Dataset for Punctuation Restoration
Sep 17, 2024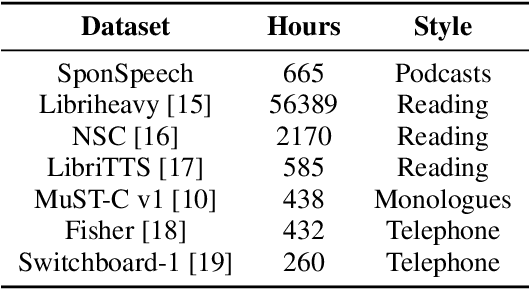

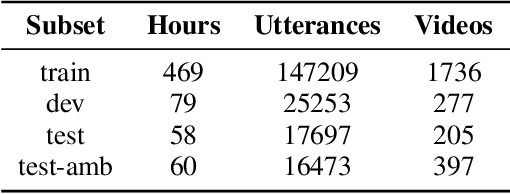
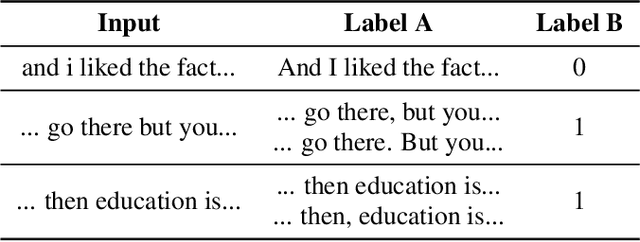
Abstract:Presently, punctuation restoration models are evaluated almost solely on well-structured, scripted corpora. On the other hand, real-world ASR systems and post-processing pipelines typically apply towards spontaneous speech with significant irregularities, stutters, and deviations from perfect grammar. To address this discrepancy, we introduce SponSpeech, a punctuation restoration dataset derived from informal speech sources, which includes punctuation and casing information. In addition to publicly releasing the dataset, we contribute a filtering pipeline that can be used to generate more data. Our filtering pipeline examines the quality of both speech audio and transcription text. We also carefully construct a ``challenging" test set, aimed at evaluating models' ability to leverage audio information to predict otherwise grammatically ambiguous punctuation. SponSpeech is available at https://github.com/GitHubAccountAnonymous/PR, along with all code for dataset building and model runs.
* 8 pages, 7 tables, 1 figure, Recognition Technologies, Inc. Technical Report
Learning Multi-view Molecular Representations with Structured and Unstructured Knowledge
Jun 14, 2024



Abstract:Capturing molecular knowledge with representation learning approaches holds significant potential in vast scientific fields such as chemistry and life science. An effective and generalizable molecular representation is expected to capture the consensus and complementary molecular expertise from diverse views and perspectives. However, existing works fall short in learning multi-view molecular representations, due to challenges in explicitly incorporating view information and handling molecular knowledge from heterogeneous sources. To address these issues, we present MV-Mol, a molecular representation learning model that harvests multi-view molecular expertise from chemical structures, unstructured knowledge from biomedical texts, and structured knowledge from knowledge graphs. We utilize text prompts to model view information and design a fusion architecture to extract view-based molecular representations. We develop a two-stage pre-training procedure, exploiting heterogeneous data of varying quality and quantity. Through extensive experiments, we show that MV-Mol provides improved representations that substantially benefit molecular property prediction. Additionally, MV-Mol exhibits state-of-the-art performance in multi-modal comprehension of molecular structures and texts. Code and data are available at https://github.com/PharMolix/OpenBioMed.
Efficient Ensemble Architecture for Multimodal Acoustic and Textual Embeddings in Punctuation Restoration using Time-Delay Neural Networks
Feb 26, 2023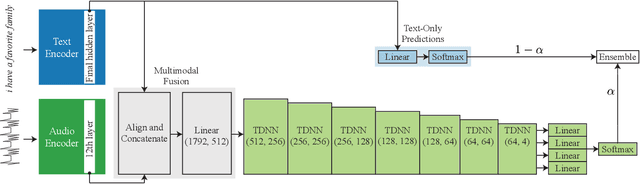
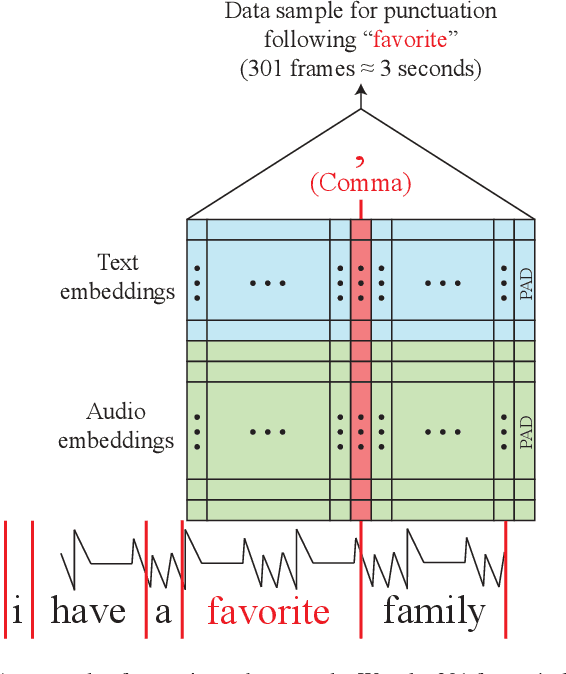


Abstract:Punctuation restoration plays an essential role in the post-processing procedure of automatic speech recognition, but model efficiency is a key requirement for this task. To that end, we present EfficientPunct, an ensemble method with a multimodal time-delay neural network that outperforms the current best model by 1.0 F1 points, using less than a tenth of its parameters to process embeddings. We streamline a speech recognizer to efficiently output hidden layer latent vectors as audio embeddings for punctuation restoration, as well as BERT to extract meaningful text embeddings. By using forced alignment and temporal convolutions, we eliminate the need for multi-head attention-based fusion, greatly increasing computational efficiency but also raising performance. EfficientPunct sets a new state of the art, in terms of both performance and efficiency, with an ensemble that weights BERT's purely language-based predictions slightly more than the multimodal network's predictions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge