Pinghong Zhou
One-shot synthesis of rare gastrointestinal lesions improves diagnostic accuracy and clinical training
Dec 30, 2025Abstract:Rare gastrointestinal lesions are infrequently encountered in routine endoscopy, restricting the data available for developing reliable artificial intelligence (AI) models and training novice clinicians. Here we present EndoRare, a one-shot, retraining-free generative framework that synthesizes diverse, high-fidelity lesion exemplars from a single reference image. By leveraging language-guided concept disentanglement, EndoRare separates pathognomonic lesion features from non-diagnostic attributes, encoding the former into a learnable prototype embedding while varying the latter to ensure diversity. We validated the framework across four rare pathologies (calcifying fibrous tumor, juvenile polyposis syndrome, familial adenomatous polyposis, and Peutz-Jeghers syndrome). Synthetic images were judged clinically plausible by experts and, when used for data augmentation, significantly enhanced downstream AI classifiers, improving the true positive rate at low false-positive rates. Crucially, a blinded reader study demonstrated that novice endoscopists exposed to EndoRare-generated cases achieved a 0.400 increase in recall and a 0.267 increase in precision. These results establish a practical, data-efficient pathway to bridge the rare-disease gap in both computer-aided diagnostics and clinical education.
EndoFinder: Online Lesion Retrieval for Explainable Colorectal Polyp Diagnosis Leveraging Latent Scene Representations
Jul 23, 2025Abstract:Colorectal cancer (CRC) remains a leading cause of cancer-related mortality, underscoring the importance of timely polyp detection and diagnosis. While deep learning models have improved optical-assisted diagnostics, they often demand extensive labeled datasets and yield "black-box" outputs with limited interpretability. In this paper, we propose EndoFinder, an online polyp retrieval framework that leverages multi-view scene representations for explainable and scalable CRC diagnosis. First, we develop a Polyp-aware Image Encoder by combining contrastive learning and a reconstruction task, guided by polyp segmentation masks. This self-supervised approach captures robust features without relying on large-scale annotated data. Next, we treat each polyp as a three-dimensional "scene" and introduce a Scene Representation Transformer, which fuses multiple views of the polyp into a single latent representation. By discretizing this representation through a hashing layer, EndoFinder enables real-time retrieval from a compiled database of historical polyp cases, where diagnostic information serves as interpretable references for new queries. We evaluate EndoFinder on both public and newly collected polyp datasets for re-identification and pathology classification. Results show that EndoFinder outperforms existing methods in accuracy while providing transparent, retrieval-based insights for clinical decision-making. By contributing a novel dataset and a scalable, explainable framework, our work addresses key challenges in polyp diagnosis and offers a promising direction for more efficient AI-driven colonoscopy workflows. The source code is available at https://github.com/ku262/EndoFinder-Scene.
Endo-CLIP: Progressive Self-Supervised Pre-training on Raw Colonoscopy Records
May 14, 2025



Abstract:Pre-training on image-text colonoscopy records offers substantial potential for improving endoscopic image analysis, but faces challenges including non-informative background images, complex medical terminology, and ambiguous multi-lesion descriptions. We introduce Endo-CLIP, a novel self-supervised framework that enhances Contrastive Language-Image Pre-training (CLIP) for this domain. Endo-CLIP's three-stage framework--cleansing, attunement, and unification--addresses these challenges by (1) removing background frames, (2) leveraging large language models to extract clinical attributes for fine-grained contrastive learning, and (3) employing patient-level cross-attention to resolve multi-polyp ambiguities. Extensive experiments demonstrate that Endo-CLIP significantly outperforms state-of-the-art pre-training methods in zero-shot and few-shot polyp detection and classification, paving the way for more accurate and clinically relevant endoscopic analysis.
Robust Polyp Detection and Diagnosis through Compositional Prompt-Guided Diffusion Models
Feb 25, 2025Abstract:Colorectal cancer (CRC) is a significant global health concern, and early detection through screening plays a critical role in reducing mortality. While deep learning models have shown promise in improving polyp detection, classification, and segmentation, their generalization across diverse clinical environments, particularly with out-of-distribution (OOD) data, remains a challenge. Multi-center datasets like PolypGen have been developed to address these issues, but their collection is costly and time-consuming. Traditional data augmentation techniques provide limited variability, failing to capture the complexity of medical images. Diffusion models have emerged as a promising solution for generating synthetic polyp images, but the image generation process in current models mainly relies on segmentation masks as the condition, limiting their ability to capture the full clinical context. To overcome these limitations, we propose a Progressive Spectrum Diffusion Model (PSDM) that integrates diverse clinical annotations-such as segmentation masks, bounding boxes, and colonoscopy reports-by transforming them into compositional prompts. These prompts are organized into coarse and fine components, allowing the model to capture both broad spatial structures and fine details, generating clinically accurate synthetic images. By augmenting training data with PSDM-generated samples, our model significantly improves polyp detection, classification, and segmentation. For instance, on the PolypGen dataset, PSDM increases the F1 score by 2.12% and the mean average precision by 3.09%, demonstrating superior performance in OOD scenarios and enhanced generalization.
EndoFinder: Online Image Retrieval for Explainable Colorectal Polyp Diagnosis
Jul 16, 2024Abstract:Determining the necessity of resecting malignant polyps during colonoscopy screen is crucial for patient outcomes, yet challenging due to the time-consuming and costly nature of histopathology examination. While deep learning-based classification models have shown promise in achieving optical biopsy with endoscopic images, they often suffer from a lack of explainability. To overcome this limitation, we introduce EndoFinder, a content-based image retrieval framework to find the 'digital twin' polyp in the reference database given a newly detected polyp. The clinical semantics of the new polyp can be inferred referring to the matched ones. EndoFinder pioneers a polyp-aware image encoder that is pre-trained on a large polyp dataset in a self-supervised way, merging masked image modeling with contrastive learning. This results in a generic embedding space ready for different downstream clinical tasks based on image retrieval. We validate the framework on polyp re-identification and optical biopsy tasks, with extensive experiments demonstrating that EndoFinder not only achieves explainable diagnostics but also matches the performance of supervised classification models. EndoFinder's reliance on image retrieval has the potential to support diverse downstream decision-making tasks during real-time colonoscopy procedures.
Knowledge Extraction and Distillation from Large-Scale Image-Text Colonoscopy Records Leveraging Large Language and Vision Models
Oct 17, 2023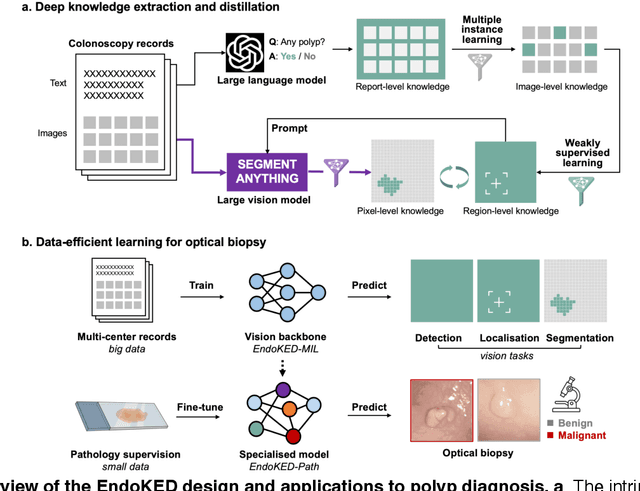
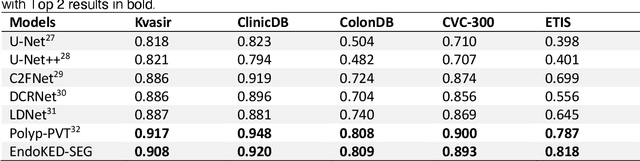
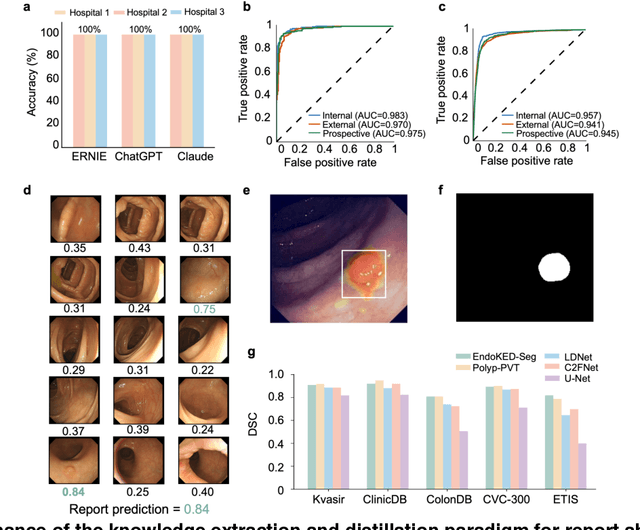
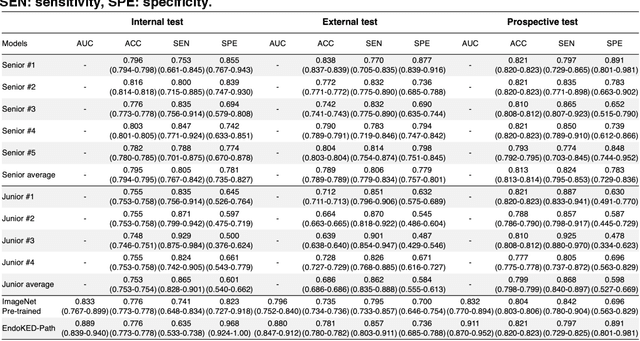
Abstract:The development of artificial intelligence systems for colonoscopy analysis often necessitates expert-annotated image datasets. However, limitations in dataset size and diversity impede model performance and generalisation. Image-text colonoscopy records from routine clinical practice, comprising millions of images and text reports, serve as a valuable data source, though annotating them is labour-intensive. Here we leverage recent advancements in large language and vision models and propose EndoKED, a data mining paradigm for deep knowledge extraction and distillation. EndoKED automates the transformation of raw colonoscopy records into image datasets with pixel-level annotation. We validate EndoKED using multi-centre datasets of raw colonoscopy records (~1 million images), demonstrating its superior performance in training polyp detection and segmentation models. Furthermore, the EndoKED pre-trained vision backbone enables data-efficient and generalisable learning for optical biopsy, achieving expert-level performance in both retrospective and prospective validation.
EndoBoost: a plug-and-play module for false positive suppression during computer-aided polyp detection in real-world colonoscopy
Dec 23, 2022Abstract:The advance of computer-aided detection systems using deep learning opened a new scope in endoscopic image analysis. However, the learning-based models developed on closed datasets are susceptible to unknown anomalies in complex clinical environments. In particular, the high false positive rate of polyp detection remains a major challenge in clinical practice. In this work, we release the FPPD-13 dataset, which provides a taxonomy and real-world cases of typical false positives during computer-aided polyp detection in real-world colonoscopy. We further propose a post-hoc module EndoBoost, which can be plugged into generic polyp detection models to filter out false positive predictions. This is realized by generative learning of the polyp manifold with normalizing flows and rejecting false positives through density estimation. Compared to supervised classification, this anomaly detection paradigm achieves better data efficiency and robustness in open-world settings. Extensive experiments demonstrate a promising false positive suppression in both retrospective and prospective validation. In addition, the released dataset can be used to perform 'stress' tests on established detection systems and encourages further research toward robust and reliable computer-aided endoscopic image analysis. The dataset and code will be publicly available at http://endoboost.miccai.cloud.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge