Xian Yang
Large-Scale Terminal Agentic Trajectory Generation from Dockerized Environments
Feb 03, 2026Abstract:Training agentic models for terminal-based tasks critically depends on high-quality terminal trajectories that capture realistic long-horizon interactions across diverse domains. However, constructing such data at scale remains challenging due to two key requirements: \textbf{\emph{Executability}}, since each instance requires a suitable and often distinct Docker environment; and \textbf{\emph{Verifiability}}, because heterogeneous task outputs preclude unified, standardized verification. To address these challenges, we propose \textbf{TerminalTraj}, a scalable pipeline that (i) filters high-quality repositories to construct Dockerized execution environments, (ii) generates Docker-aligned task instances, and (iii) synthesizes agent trajectories with executable validation code. Using TerminalTraj, we curate 32K Docker images and generate 50,733 verified terminal trajectories across eight domains. Models trained on this data with the Qwen2.5-Coder backbone achieve consistent performance improvements on TerminalBench (TB), with gains of up to 20\% on TB~1.0 and 10\% on TB~2.0 over their respective backbones. Notably, \textbf{TerminalTraj-32B} achieves strong performance among models with fewer than 100B parameters, reaching 35.30\% on TB~1.0 and 22.00\% on TB~2.0, and demonstrates improved test-time scaling behavior. All code and data are available at https://github.com/Wusiwei0410/TerminalTraj.
One-shot synthesis of rare gastrointestinal lesions improves diagnostic accuracy and clinical training
Dec 30, 2025Abstract:Rare gastrointestinal lesions are infrequently encountered in routine endoscopy, restricting the data available for developing reliable artificial intelligence (AI) models and training novice clinicians. Here we present EndoRare, a one-shot, retraining-free generative framework that synthesizes diverse, high-fidelity lesion exemplars from a single reference image. By leveraging language-guided concept disentanglement, EndoRare separates pathognomonic lesion features from non-diagnostic attributes, encoding the former into a learnable prototype embedding while varying the latter to ensure diversity. We validated the framework across four rare pathologies (calcifying fibrous tumor, juvenile polyposis syndrome, familial adenomatous polyposis, and Peutz-Jeghers syndrome). Synthetic images were judged clinically plausible by experts and, when used for data augmentation, significantly enhanced downstream AI classifiers, improving the true positive rate at low false-positive rates. Crucially, a blinded reader study demonstrated that novice endoscopists exposed to EndoRare-generated cases achieved a 0.400 increase in recall and a 0.267 increase in precision. These results establish a practical, data-efficient pathway to bridge the rare-disease gap in both computer-aided diagnostics and clinical education.
Shared Object Manipulation with a Team of Collaborative Quadrupeds
Oct 01, 2025
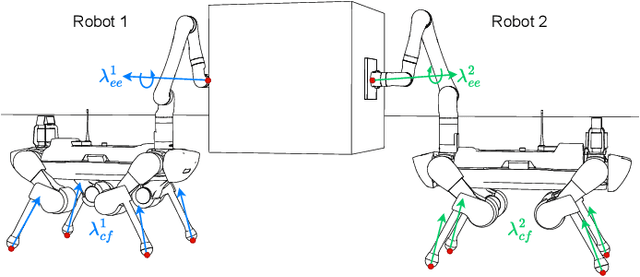
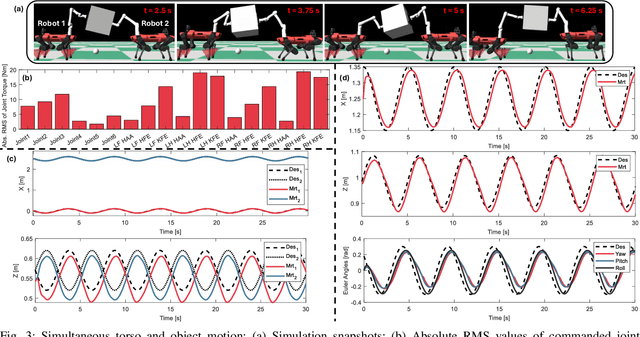
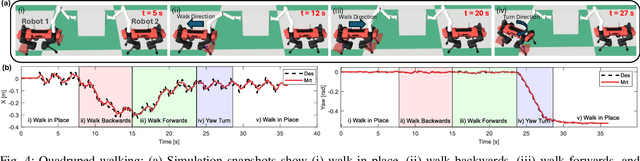
Abstract:Utilizing teams of multiple robots is advantageous for handling bulky objects. Many related works focus on multi-manipulator systems, which are limited by workspace constraints. In this paper, we extend a classical hybrid motion-force controller to a team of legged manipulator systems, enabling collaborative loco-manipulation of rigid objects with a force-closed grasp. Our novel approach allows the robots to flexibly coordinate their movements, achieving efficient and stable object co-manipulation and transport, validated through extensive simulations and real-world experiments.
EndoFinder: Online Lesion Retrieval for Explainable Colorectal Polyp Diagnosis Leveraging Latent Scene Representations
Jul 23, 2025Abstract:Colorectal cancer (CRC) remains a leading cause of cancer-related mortality, underscoring the importance of timely polyp detection and diagnosis. While deep learning models have improved optical-assisted diagnostics, they often demand extensive labeled datasets and yield "black-box" outputs with limited interpretability. In this paper, we propose EndoFinder, an online polyp retrieval framework that leverages multi-view scene representations for explainable and scalable CRC diagnosis. First, we develop a Polyp-aware Image Encoder by combining contrastive learning and a reconstruction task, guided by polyp segmentation masks. This self-supervised approach captures robust features without relying on large-scale annotated data. Next, we treat each polyp as a three-dimensional "scene" and introduce a Scene Representation Transformer, which fuses multiple views of the polyp into a single latent representation. By discretizing this representation through a hashing layer, EndoFinder enables real-time retrieval from a compiled database of historical polyp cases, where diagnostic information serves as interpretable references for new queries. We evaluate EndoFinder on both public and newly collected polyp datasets for re-identification and pathology classification. Results show that EndoFinder outperforms existing methods in accuracy while providing transparent, retrieval-based insights for clinical decision-making. By contributing a novel dataset and a scalable, explainable framework, our work addresses key challenges in polyp diagnosis and offers a promising direction for more efficient AI-driven colonoscopy workflows. The source code is available at https://github.com/ku262/EndoFinder-Scene.
Endo-CLIP: Progressive Self-Supervised Pre-training on Raw Colonoscopy Records
May 14, 2025



Abstract:Pre-training on image-text colonoscopy records offers substantial potential for improving endoscopic image analysis, but faces challenges including non-informative background images, complex medical terminology, and ambiguous multi-lesion descriptions. We introduce Endo-CLIP, a novel self-supervised framework that enhances Contrastive Language-Image Pre-training (CLIP) for this domain. Endo-CLIP's three-stage framework--cleansing, attunement, and unification--addresses these challenges by (1) removing background frames, (2) leveraging large language models to extract clinical attributes for fine-grained contrastive learning, and (3) employing patient-level cross-attention to resolve multi-polyp ambiguities. Extensive experiments demonstrate that Endo-CLIP significantly outperforms state-of-the-art pre-training methods in zero-shot and few-shot polyp detection and classification, paving the way for more accurate and clinically relevant endoscopic analysis.
Robust Polyp Detection and Diagnosis through Compositional Prompt-Guided Diffusion Models
Feb 25, 2025Abstract:Colorectal cancer (CRC) is a significant global health concern, and early detection through screening plays a critical role in reducing mortality. While deep learning models have shown promise in improving polyp detection, classification, and segmentation, their generalization across diverse clinical environments, particularly with out-of-distribution (OOD) data, remains a challenge. Multi-center datasets like PolypGen have been developed to address these issues, but their collection is costly and time-consuming. Traditional data augmentation techniques provide limited variability, failing to capture the complexity of medical images. Diffusion models have emerged as a promising solution for generating synthetic polyp images, but the image generation process in current models mainly relies on segmentation masks as the condition, limiting their ability to capture the full clinical context. To overcome these limitations, we propose a Progressive Spectrum Diffusion Model (PSDM) that integrates diverse clinical annotations-such as segmentation masks, bounding boxes, and colonoscopy reports-by transforming them into compositional prompts. These prompts are organized into coarse and fine components, allowing the model to capture both broad spatial structures and fine details, generating clinically accurate synthetic images. By augmenting training data with PSDM-generated samples, our model significantly improves polyp detection, classification, and segmentation. For instance, on the PolypGen dataset, PSDM increases the F1 score by 2.12% and the mean average precision by 3.09%, demonstrating superior performance in OOD scenarios and enhanced generalization.
C-LoRA: Continual Low-Rank Adaptation for Pre-trained Models
Feb 25, 2025



Abstract:Low-Rank Adaptation (LoRA) is an efficient fine-tuning method that has been extensively applied in areas such as natural language processing and computer vision. Existing LoRA fine-tuning approaches excel in static environments but struggle in dynamic learning due to reliance on multiple adapter modules, increasing overhead and complicating inference. We propose Continual Low-Rank Adaptation (C-LoRA), a novel extension of LoRA for continual learning. C-LoRA uses a learnable routing matrix to dynamically manage parameter updates across tasks, ensuring efficient reuse of learned subspaces while enforcing orthogonality to minimize interference and forgetting. Unlike existing approaches that require separate adapters for each task, C-LoRA enables a integrated approach for task adaptation, achieving both scalability and parameter efficiency in sequential learning scenarios. C-LoRA achieves state-of-the-art accuracy and parameter efficiency on benchmarks while providing theoretical insights into its routing matrix's role in retaining and transferring knowledge, establishing a scalable framework for continual learning.
Progressive Local Alignment for Medical Multimodal Pre-training
Feb 25, 2025Abstract:Local alignment between medical images and text is essential for accurate diagnosis, though it remains challenging due to the absence of natural local pairings and the limitations of rigid region recognition methods. Traditional approaches rely on hard boundaries, which introduce uncertainty, whereas medical imaging demands flexible soft region recognition to handle irregular structures. To overcome these challenges, we propose the Progressive Local Alignment Network (PLAN), which designs a novel contrastive learning-based approach for local alignment to establish meaningful word-pixel relationships and introduces a progressive learning strategy to iteratively refine these relationships, enhancing alignment precision and robustness. By combining these techniques, PLAN effectively improves soft region recognition while suppressing noise interference. Extensive experiments on multiple medical datasets demonstrate that PLAN surpasses state-of-the-art methods in phrase grounding, image-text retrieval, object detection, and zero-shot classification, setting a new benchmark for medical image-text alignment.
Unlocking Multimodal Integration in EHRs: A Prompt Learning Framework for Language and Time Series Fusion
Feb 19, 2025Abstract:Large language models (LLMs) have shown remarkable performance in vision-language tasks, but their application in the medical field remains underexplored, particularly for integrating structured time series data with unstructured clinical notes. In clinical practice, dynamic time series data such as lab test results capture critical temporal patterns, while clinical notes provide rich semantic context. Merging these modalities is challenging due to the inherent differences between continuous signals and discrete text. To bridge this gap, we introduce ProMedTS, a novel self-supervised multimodal framework that employs prompt-guided learning to unify these heterogeneous data types. Our approach leverages lightweight anomaly detection to generate anomaly captions that serve as prompts, guiding the encoding of raw time series data into informative embeddings. These embeddings are aligned with textual representations in a shared latent space, preserving fine-grained temporal nuances alongside semantic insights. Furthermore, our framework incorporates tailored self-supervised objectives to enhance both intra- and inter-modal alignment. We evaluate ProMedTS on disease diagnosis tasks using real-world datasets, and the results demonstrate that our method consistently outperforms state-of-the-art approaches.
MinMo: A Multimodal Large Language Model for Seamless Voice Interaction
Jan 10, 2025



Abstract:Recent advancements in large language models (LLMs) and multimodal speech-text models have laid the groundwork for seamless voice interactions, enabling real-time, natural, and human-like conversations. Previous models for voice interactions are categorized as native and aligned. Native models integrate speech and text processing in one framework but struggle with issues like differing sequence lengths and insufficient pre-training. Aligned models maintain text LLM capabilities but are often limited by small datasets and a narrow focus on speech tasks. In this work, we introduce MinMo, a Multimodal Large Language Model with approximately 8B parameters for seamless voice interaction. We address the main limitations of prior aligned multimodal models. We train MinMo through multiple stages of speech-to-text alignment, text-to-speech alignment, speech-to-speech alignment, and duplex interaction alignment, on 1.4 million hours of diverse speech data and a broad range of speech tasks. After the multi-stage training, MinMo achieves state-of-the-art performance across various benchmarks for voice comprehension and generation while maintaining the capabilities of text LLMs, and also facilitates full-duplex conversation, that is, simultaneous two-way communication between the user and the system. Moreover, we propose a novel and simple voice decoder that outperforms prior models in voice generation. The enhanced instruction-following capabilities of MinMo supports controlling speech generation based on user instructions, with various nuances including emotions, dialects, and speaking rates, and mimicking specific voices. For MinMo, the speech-to-text latency is approximately 100ms, full-duplex latency is approximately 600ms in theory and 800ms in practice. The MinMo project web page is https://funaudiollm.github.io/minmo, and the code and models will be released soon.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge