Shuai Niu
Unlocking Multimodal Integration in EHRs: A Prompt Learning Framework for Language and Time Series Fusion
Feb 19, 2025Abstract:Large language models (LLMs) have shown remarkable performance in vision-language tasks, but their application in the medical field remains underexplored, particularly for integrating structured time series data with unstructured clinical notes. In clinical practice, dynamic time series data such as lab test results capture critical temporal patterns, while clinical notes provide rich semantic context. Merging these modalities is challenging due to the inherent differences between continuous signals and discrete text. To bridge this gap, we introduce ProMedTS, a novel self-supervised multimodal framework that employs prompt-guided learning to unify these heterogeneous data types. Our approach leverages lightweight anomaly detection to generate anomaly captions that serve as prompts, guiding the encoding of raw time series data into informative embeddings. These embeddings are aligned with textual representations in a shared latent space, preserving fine-grained temporal nuances alongside semantic insights. Furthermore, our framework incorporates tailored self-supervised objectives to enhance both intra- and inter-modal alignment. We evaluate ProMedTS on disease diagnosis tasks using real-world datasets, and the results demonstrate that our method consistently outperforms state-of-the-art approaches.
Multimodal Clinical Reasoning through Knowledge-augmented Rationale Generation
Nov 12, 2024Abstract:Clinical rationales play a pivotal role in accurate disease diagnosis; however, many models predominantly use discriminative methods and overlook the importance of generating supportive rationales. Rationale distillation is a process that transfers knowledge from large language models (LLMs) to smaller language models (SLMs), thereby enhancing the latter's ability to break down complex tasks. Despite its benefits, rationale distillation alone is inadequate for addressing domain knowledge limitations in tasks requiring specialized expertise, such as disease diagnosis. Effectively embedding domain knowledge in SLMs poses a significant challenge. While current LLMs are primarily geared toward processing textual data, multimodal LLMs that incorporate time series data, especially electronic health records (EHRs), are still evolving. To tackle these limitations, we introduce ClinRaGen, an SLM optimized for multimodal rationale generation in disease diagnosis. ClinRaGen incorporates a unique knowledge-augmented attention mechanism to merge domain knowledge with time series EHR data, utilizing a stepwise rationale distillation strategy to produce both textual and time series-based clinical rationales. Our evaluations show that ClinRaGen markedly improves the SLM's capability to interpret multimodal EHR data and generate accurate clinical rationales, supporting more reliable disease diagnosis, advancing LLM applications in healthcare, and narrowing the performance divide between LLMs and SLMs.
Label-dependent and event-guided interpretable disease risk prediction using EHRs
Jan 18, 2022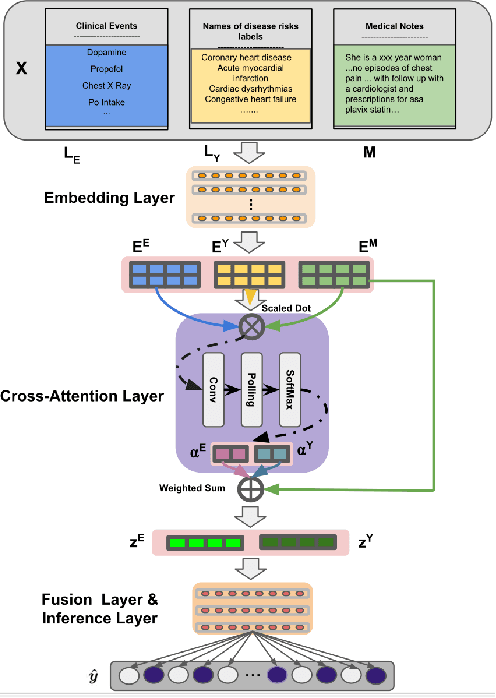
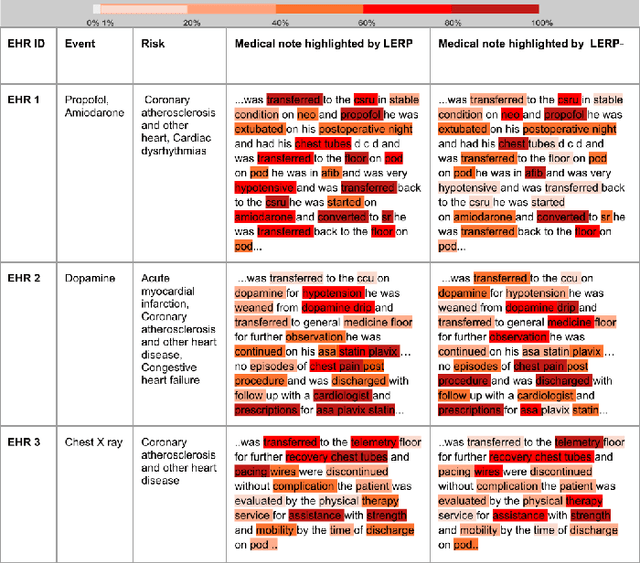
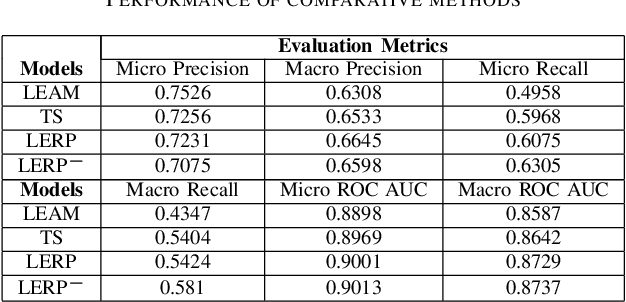
Abstract:Electronic health records (EHRs) contain patients' heterogeneous data that are collected from medical providers involved in the patient's care, including medical notes, clinical events, laboratory test results, symptoms, and diagnoses. In the field of modern healthcare, predicting whether patients would experience any risks based on their EHRs has emerged as a promising research area, in which artificial intelligence (AI) plays a key role. To make AI models practically applicable, it is required that the prediction results should be both accurate and interpretable. To achieve this goal, this paper proposed a label-dependent and event-guided risk prediction model (LERP) to predict the presence of multiple disease risks by mainly extracting information from unstructured medical notes. Our model is featured in the following aspects. First, we adopt a label-dependent mechanism that gives greater attention to words from medical notes that are semantically similar to the names of risk labels. Secondly, as the clinical events (e.g., treatments and drugs) can also indicate the health status of patients, our model utilizes the information from events and uses them to generate an event-guided representation of medical notes. Thirdly, both label-dependent and event-guided representations are integrated to make a robust prediction, in which the interpretability is enabled by the attention weights over words from medical notes. To demonstrate the applicability of the proposed method, we apply it to the MIMIC-III dataset, which contains real-world EHRs collected from hospitals. Our method is evaluated in both quantitative and qualitative ways.
Label Dependent Attention Model for Disease Risk Prediction Using Multimodal Electronic Health Records
Jan 18, 2022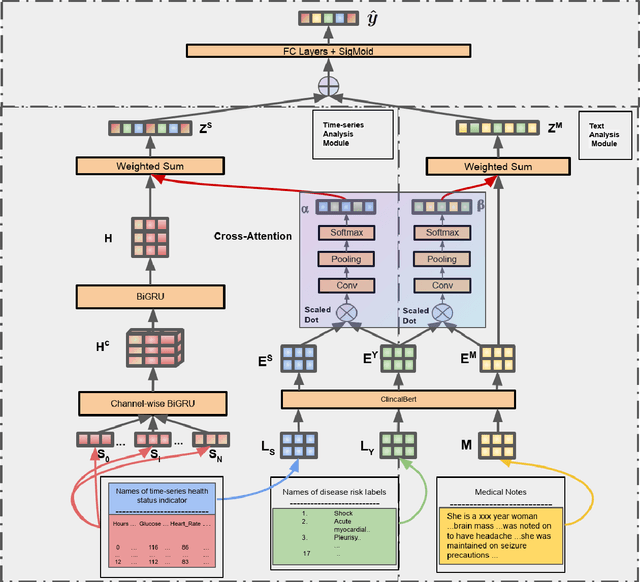
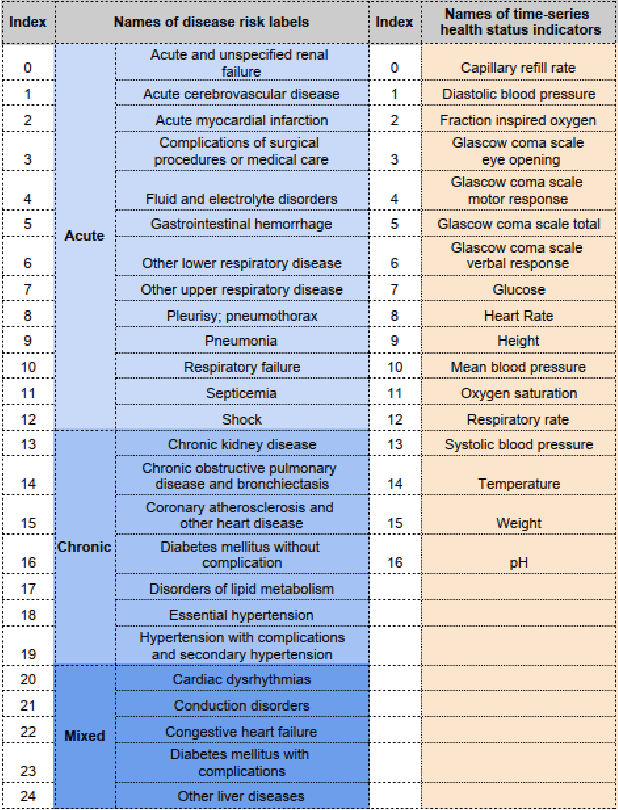
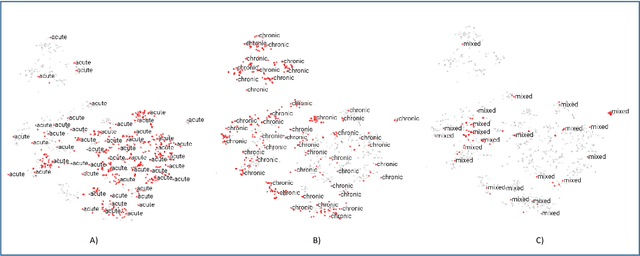
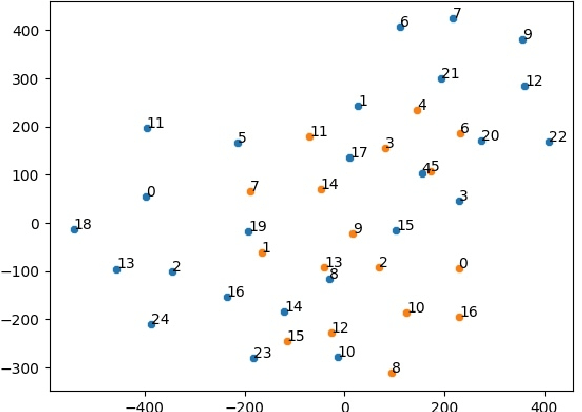
Abstract:Disease risk prediction has attracted increasing attention in the field of modern healthcare, especially with the latest advances in artificial intelligence (AI). Electronic health records (EHRs), which contain heterogeneous patient information, are widely used in disease risk prediction tasks. One challenge of applying AI models for risk prediction lies in generating interpretable evidence to support the prediction results while retaining the prediction ability. In order to address this problem, we propose the method of jointly embedding words and labels whereby attention modules learn the weights of words from medical notes according to their relevance to the names of risk prediction labels. This approach boosts interpretability by employing an attention mechanism and including the names of prediction tasks in the model. However, its application is only limited to the handling of textual inputs such as medical notes. In this paper, we propose a label dependent attention model LDAM to 1) improve the interpretability by exploiting Clinical-BERT (a biomedical language model pre-trained on a large clinical corpus) to encode biomedically meaningful features and labels jointly; 2) extend the idea of joint embedding to the processing of time-series data, and develop a multi-modal learning framework for integrating heterogeneous information from medical notes and time-series health status indicators. To demonstrate our method, we apply LDAM to the MIMIC-III dataset to predict different disease risks. We evaluate our method both quantitatively and qualitatively. Specifically, the predictive power of LDAM will be shown, and case studies will be carried out to illustrate its interpretability.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge