Karina Zadorozhny
Similarity-Quantized Relative Difference Learning for Improved Molecular Activity Prediction
Jan 15, 2025



Abstract:Accurate prediction of molecular activities is crucial for efficient drug discovery, yet remains challenging due to limited and noisy datasets. We introduce Similarity-Quantized Relative Learning (SQRL), a learning framework that reformulates molecular activity prediction as relative difference learning between structurally similar pairs of compounds. SQRL uses precomputed molecular similarities to enhance training of graph neural networks and other architectures, and significantly improves accuracy and generalization in low-data regimes common in drug discovery. We demonstrate its broad applicability and real-world potential through benchmarking on public datasets as well as proprietary industry data. Our findings demonstrate that leveraging similarity-aware relative differences provides an effective paradigm for molecular activity prediction.
Protein Discovery with Discrete Walk-Jump Sampling
Jun 08, 2023



Abstract:We resolve difficulties in training and sampling from a discrete generative model by learning a smoothed energy function, sampling from the smoothed data manifold with Langevin Markov chain Monte Carlo (MCMC), and projecting back to the true data manifold with one-step denoising. Our Discrete Walk-Jump Sampling formalism combines the maximum likelihood training of an energy-based model and improved sample quality of a score-based model, while simplifying training and sampling by requiring only a single noise level. We evaluate the robustness of our approach on generative modeling of antibody proteins and introduce the distributional conformity score to benchmark protein generative models. By optimizing and sampling from our models for the proposed distributional conformity score, 97-100% of generated samples are successfully expressed and purified and 35% of functional designs show equal or improved binding affinity compared to known functional antibodies on the first attempt in a single round of laboratory experiments. We also report the first demonstration of long-run fast-mixing MCMC chains where diverse antibody protein classes are visited in a single MCMC chain.
Out-of-Distribution Detection for Medical Applications: Guidelines for Practical Evaluation
Sep 30, 2021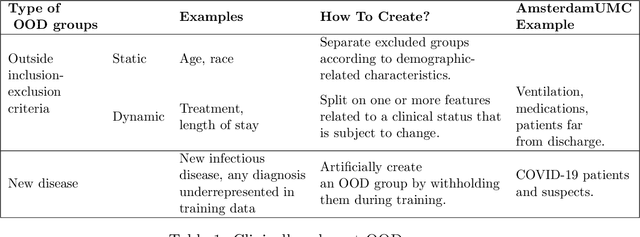
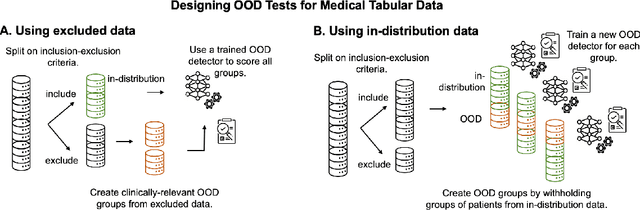
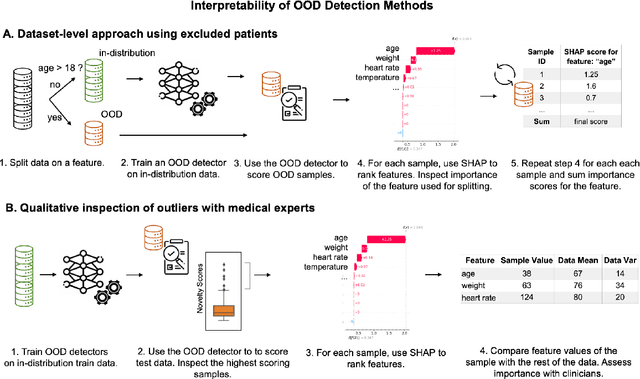
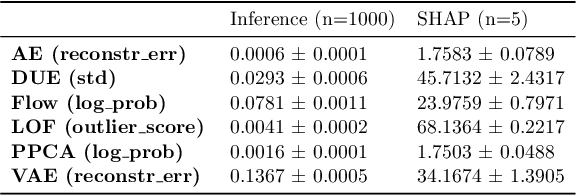
Abstract:Detection of Out-of-Distribution (OOD) samples in real time is a crucial safety check for deployment of machine learning models in the medical field. Despite a growing number of uncertainty quantification techniques, there is a lack of evaluation guidelines on how to select OOD detection methods in practice. This gap impedes implementation of OOD detection methods for real-world applications. Here, we propose a series of practical considerations and tests to choose the best OOD detector for a specific medical dataset. These guidelines are illustrated on a real-life use case of Electronic Health Records (EHR). Our results can serve as a guide for implementation of OOD detection methods in clinical practice, mitigating risks associated with the use of machine learning models in healthcare.
Deep Denerative Models for Drug Design and Response
Sep 14, 2021



Abstract:Designing new chemical compounds with desired pharmaceutical properties is a challenging task and takes years of development and testing. Still, a majority of new drugs fail to prove efficient. Recent success of deep generative modeling holds promises of generation and optimization of new molecules. In this review paper, we provide an overview of the current generative models, and describe necessary biological and chemical terminology, including molecular representations needed to understand the field of drug design and drug response. We present commonly used chemical and biological databases, and tools for generative modeling. Finally, we summarize the current state of generative modeling for drug design and drug response prediction, highlighting the state-of-art approaches and limitations the field is currently facing.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge