Anna Bogdanova
Data Collaboration Analysis applied to Compound Datasets and the Introduction of Projection data to Non-IID settings
Aug 01, 2023Abstract:Given the time and expense associated with bringing a drug to market, numerous studies have been conducted to predict the properties of compounds based on their structure using machine learning. Federated learning has been applied to compound datasets to increase their prediction accuracy while safeguarding potentially proprietary information. However, federated learning is encumbered by low accuracy in not identically and independently distributed (non-IID) settings, i.e., data partitioning has a large label bias, and is considered unsuitable for compound datasets, which tend to have large label bias. To address this limitation, we utilized an alternative method of distributed machine learning to chemical compound data from open sources, called data collaboration analysis (DC). We also proposed data collaboration analysis using projection data (DCPd), which is an improved method that utilizes auxiliary PubChem data. This improves the quality of individual user-side data transformations for the projection data for the creation of intermediate representations. The classification accuracy, i.e., area under the curve in the receiver operating characteristic curve (ROC-AUC) and AUC in the precision-recall curve (PR-AUC), of federated averaging (FedAvg), DC, and DCPd was compared for five compound datasets. We determined that the machine learning performance for non-IID settings was in the order of DCPd, DC, and FedAvg, although they were almost the same in identically and independently distributed (IID) settings. Moreover, the results showed that compared to other methods, DCPd exhibited a negligible decline in classification accuracy in experiments with different degrees of label bias. Thus, DCPd can address the low performance in non-IID settings, which is one of the challenges of federated learning.
Topologically-Regularized Multiple Instance Learning for Red Blood Cell Disease Classification
Jul 26, 2023Abstract:Diagnosing rare anemia disorders using microscopic images is challenging for skilled specialists and machine-learning methods alike. Due to thousands of disease-relevant cells in a single blood sample, this constitutes a complex multiple-instance learning (MIL) problem. While the spatial neighborhood of red blood cells is not meaningful per se, the topology, i.e., the geometry of blood samples as a whole, contains informative features to remedy typical MIL issues, such as vanishing gradients and overfitting when training on limited data. We thus develop a topology-based approach that extracts multi-scale topological features from bags of single red blood cell images. The topological features are used to regularize the model, enforcing the preservation of characteristic topological properties of the data. Applied to a dataset of 71 patients suffering from rare anemia disorders with 521 microscopic images of red blood cells, our experiments show that topological regularization is an effective method that leads to more than 3% performance improvements for the automated classification of rare anemia disorders based on single-cell images. This is the first approach that uses topological properties for regularizing the MIL process.
Achieving Transparency in Distributed Machine Learning with Explainable Data Collaboration
Dec 06, 2022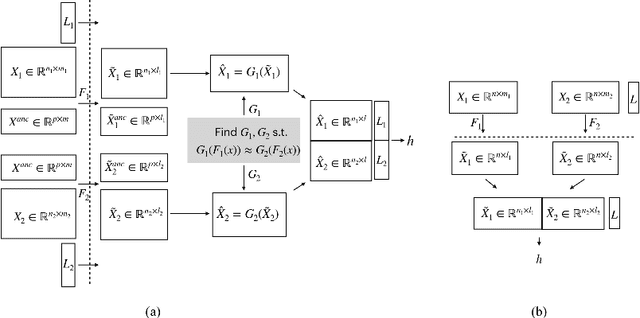
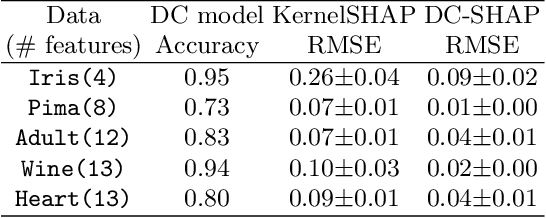
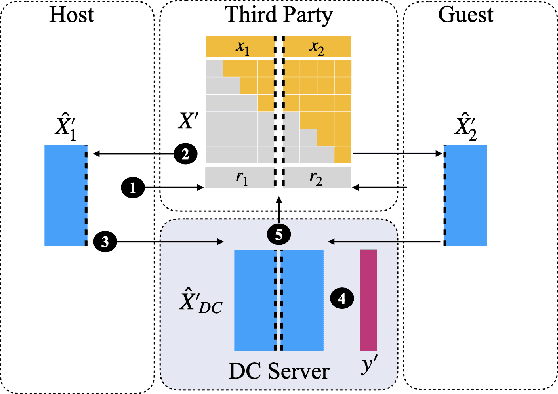
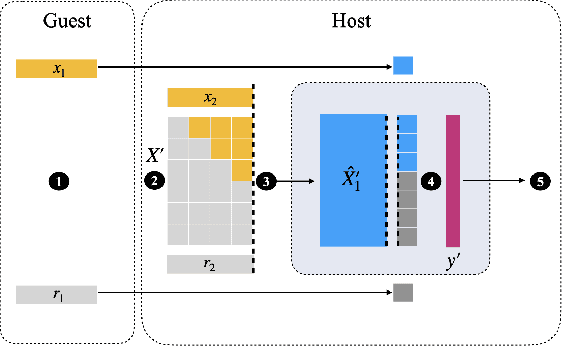
Abstract:Transparency of Machine Learning models used for decision support in various industries becomes essential for ensuring their ethical use. To that end, feature attribution methods such as SHAP (SHapley Additive exPlanations) are widely used to explain the predictions of black-box machine learning models to customers and developers. However, a parallel trend has been to train machine learning models in collaboration with other data holders without accessing their data. Such models, trained over horizontally or vertically partitioned data, present a challenge for explainable AI because the explaining party may have a biased view of background data or a partial view of the feature space. As a result, explanations obtained from different participants of distributed machine learning might not be consistent with one another, undermining trust in the product. This paper presents an Explainable Data Collaboration Framework based on a model-agnostic additive feature attribution algorithm (KernelSHAP) and Data Collaboration method of privacy-preserving distributed machine learning. In particular, we present three algorithms for different scenarios of explainability in Data Collaboration and verify their consistency with experiments on open-access datasets. Our results demonstrated a significant (by at least a factor of 1.75) decrease in feature attribution discrepancies among the users of distributed machine learning.
Anomaly-aware multiple instance learning for rare anemia disorder classification
Jul 04, 2022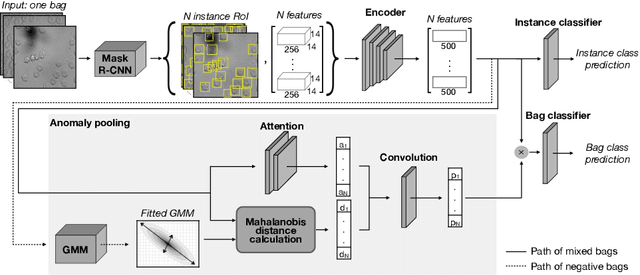

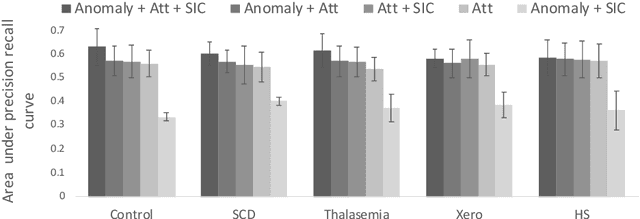
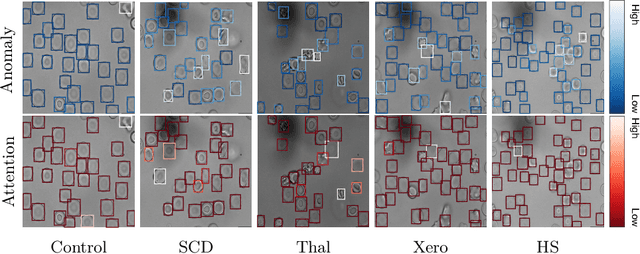
Abstract:Deep learning-based classification of rare anemia disorders is challenged by the lack of training data and instance-level annotations. Multiple Instance Learning (MIL) has shown to be an effective solution, yet it suffers from low accuracy and limited explainability. Although the inclusion of attention mechanisms has addressed these issues, their effectiveness highly depends on the amount and diversity of cells in the training samples. Consequently, the poor machine learning performance on rare anemia disorder classification from blood samples remains unresolved. In this paper, we propose an interpretable pooling method for MIL to address these limitations. By benefiting from instance-level information of negative bags (i.e., homogeneous benign cells from healthy individuals), our approach increases the contribution of anomalous instances. We show that our strategy outperforms standard MIL classification algorithms and provides a meaningful explanation behind its decisions. Moreover, it can denote anomalous instances of rare blood diseases that are not seen during the training phase.
Sickle Cell Disease Severity Prediction from Percoll Gradient Images using Graph Convolutional Networks
Sep 11, 2021
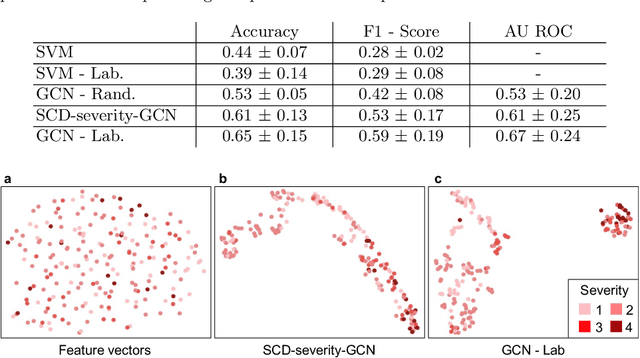

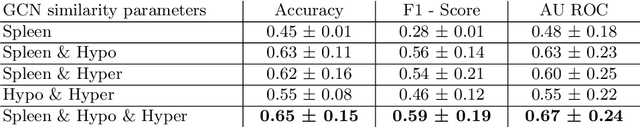
Abstract:Sickle cell disease (SCD) is a severe genetic hemoglobin disorder that results in premature destruction of red blood cells. Assessment of the severity of the disease is a challenging task in clinical routine since the causes of broad variance in SCD manifestation despite the common genetic cause remain unclear. Identification of the biomarkers that would predict the severity grade is of importance for prognosis and assessment of responsiveness of patients to therapy. Detection of the changes in red blood cell (RBC) density through separation of Percoll density gradient could be such marker as it allows to resolve intercellular differences and follow the most damaged dense cells prone to destruction and vaso-occlusion. Quantification of the images obtained from the distribution of RBCs in Percoll gradient and interpretation of the obtained is an important prerequisite for establishment of this approach. Here, we propose a novel approach combining a graph convolutional network, a convolutional neural network, fast Fourier transform, and recursive feature elimination to predict the severity of SCD directly from a Percoll image. Two important but expensive laboratory blood test parameters measurements are used for training the graph convolutional network. To make the model independent from such tests during prediction, the two parameters are estimated by a neural network from the Percoll image directly. On a cohort of 216 subjects, we achieve a prediction performance that is only slightly below an approach where the groundtruth laboratory measurements are used. Our proposed method is the first computational approach for the difficult task of SCD severity prediction. The two-step approach relies solely on inexpensive and simple blood analysis tools and can have a significant impact on the patients' survival in underdeveloped countries where access to medical instruments and doctors is limited
Fourier Transform of Percoll Gradients Boosts CNN Classification of Hereditary Hemolytic Anemias
Mar 17, 2021

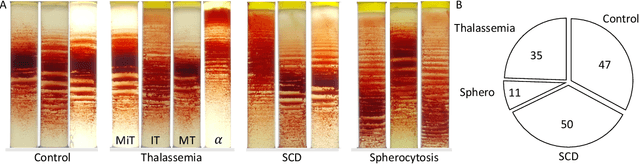
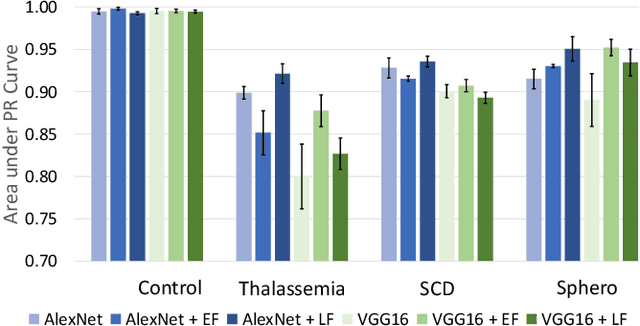
Abstract:Hereditary hemolytic anemias are genetic disorders that affect the shape and density of red blood cells. Genetic tests currently used to diagnose such anemias are expensive and unavailable in the majority of clinical labs. Here, we propose a method for identifying hereditary hemolytic anemias based on a standard biochemistry method, called Percoll gradient, obtained by centrifuging a patient's blood. Our hybrid approach consists on using spatial data-driven features, extracted with a convolutional neural network and spectral handcrafted features obtained from fast Fourier transform. We compare late and early feature fusion with AlexNet and VGG16 architectures. AlexNet with late fusion of spectral features performs better compared to other approaches. We achieved an average F1-score of 88% on different classes suggesting the possibility of diagnosing of hereditary hemolytic anemias from Percoll gradients. Finally, we utilize Grad-CAM to explore the spatial features used for classification.
Accuracy and Privacy Evaluations of Collaborative Data Analysis
Jan 27, 2021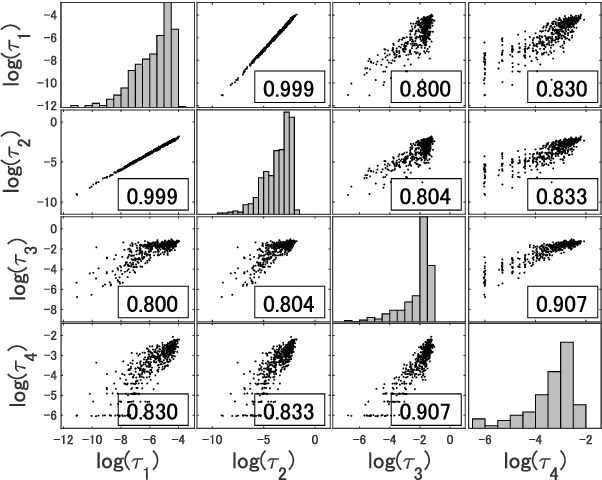
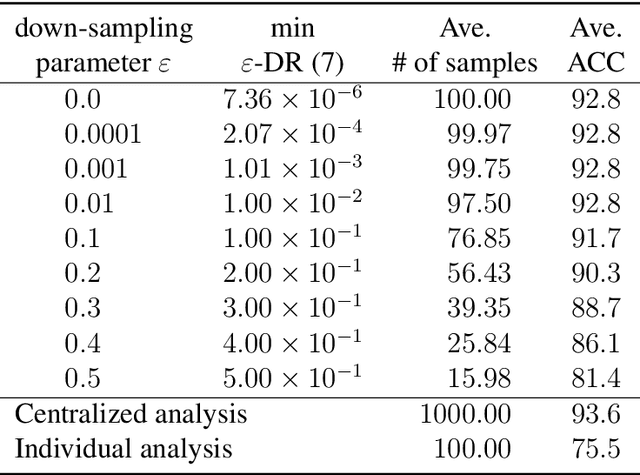
Abstract:Distributed data analysis without revealing the individual data has recently attracted significant attention in several applications. A collaborative data analysis through sharing dimensionality reduced representations of data has been proposed as a non-model sharing-type federated learning. This paper analyzes the accuracy and privacy evaluations of this novel framework. In the accuracy analysis, we provided sufficient conditions for the equivalence of the collaborative data analysis and the centralized analysis with dimensionality reduction. In the privacy analysis, we proved that collaborative users' private datasets are protected with a double privacy layer against insider and external attacking scenarios.
* 16 pages; 2 figures; 1 table
Federated Learning System without Model Sharing through Integration of Dimensional Reduced Data Representations
Nov 13, 2020
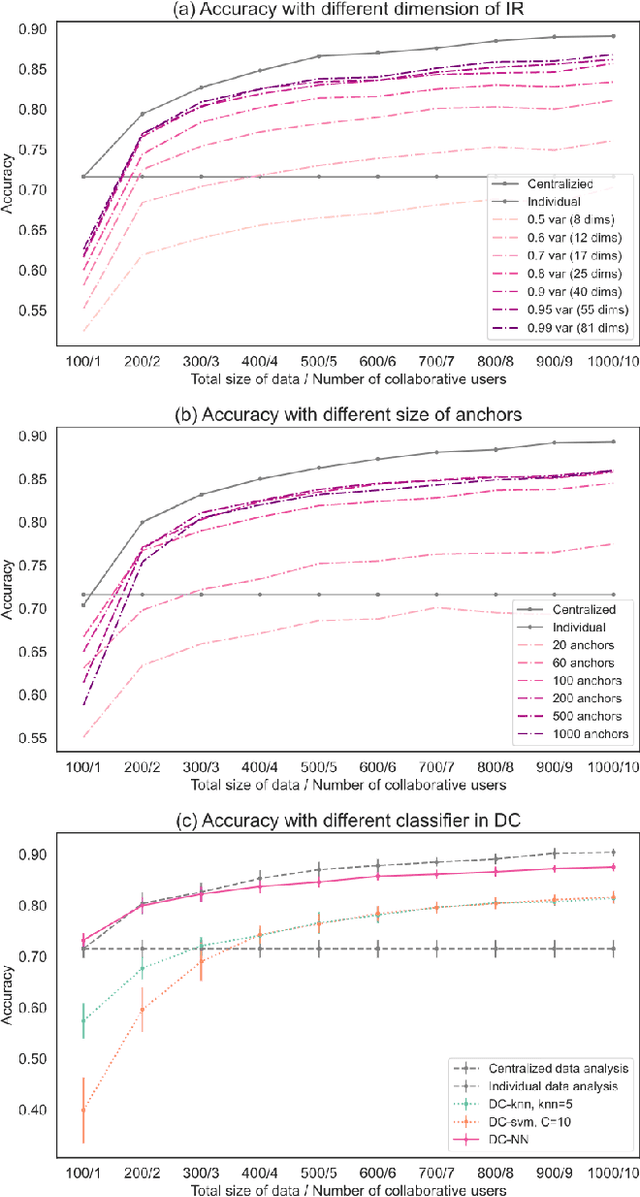
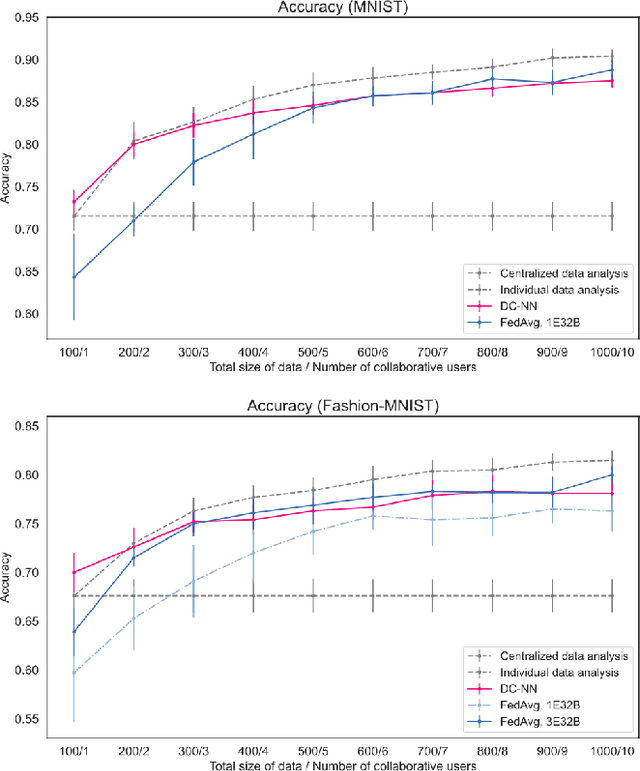
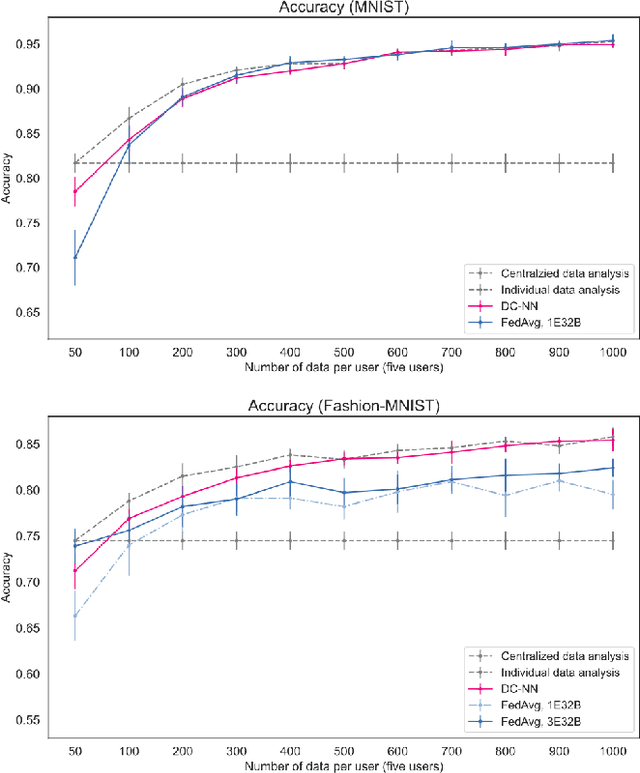
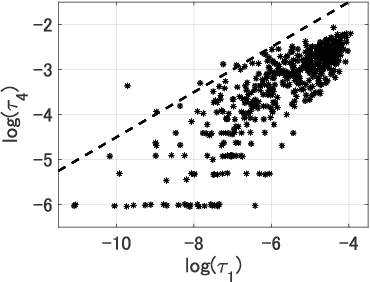
Abstract:Dimensionality Reduction is a commonly used element in a machine learning pipeline that helps to extract important features from high-dimensional data. In this work, we explore an alternative federated learning system that enables integration of dimensionality reduced representations of distributed data prior to a supervised learning task, thus avoiding model sharing among the parties. We compare the performance of this approach on image classification tasks to three alternative frameworks: centralized machine learning, individual machine learning, and Federated Averaging, and analyze potential use cases for a federated learning system without model sharing. Our results show that our approach can achieve similar accuracy as Federated Averaging and performs better than Federated Averaging in a small-user setting.
Attention based Multiple Instance Learning for Classification of Blood Cell Disorders
Jul 22, 2020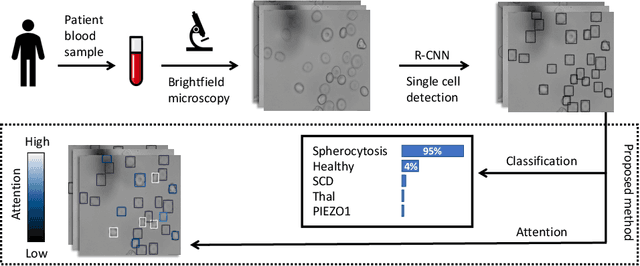
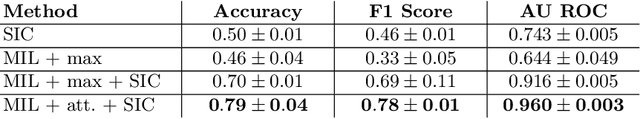
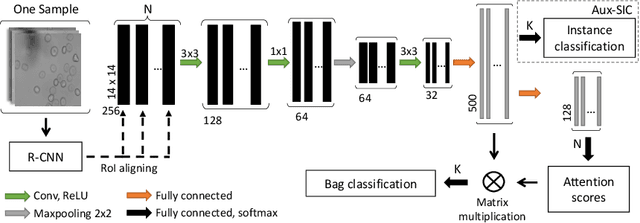
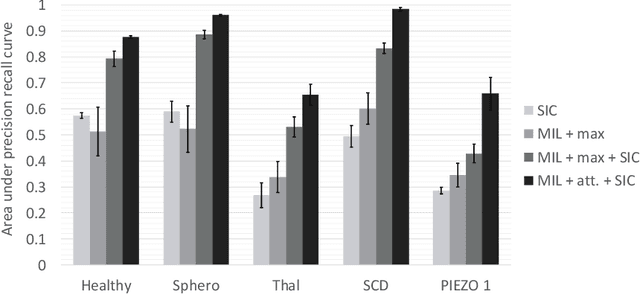
Abstract:Red blood cells are highly deformable and present in various shapes. In blood cell disorders, only a subset of all cells is morphologically altered and relevant for the diagnosis. However, manually labeling of all cells is laborious, complicated and introduces inter-expert variability. We propose an attention based multiple instance learning method to classify blood samples of patients suffering from blood cell disorders. Cells are detected using an R-CNN architecture. With the features extracted for each cell, a multiple instance learning method classifies patient samples into one out of four blood cell disorders. The attention mechanism provides a measure of the contribution of each cell to the overall classification and significantly improves the network's classification accuracy as well as its interpretability for the medical expert.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge