Hiroyuki Abe
Domain Generalization of Pathological Image Segmentation by Patch-Level and WSI-Level Contrastive Learning
Aug 11, 2025


Abstract:In this paper, we address domain shifts in pathological images by focusing on shifts within whole slide images~(WSIs), such as patient characteristics and tissue thickness, rather than shifts between hospitals. Traditional approaches rely on multi-hospital data, but data collection challenges often make this impractical. Therefore, the proposed domain generalization method captures and leverages intra-hospital domain shifts by clustering WSI-level features from non-tumor regions and treating these clusters as domains. To mitigate domain shift, we apply contrastive learning to reduce feature gaps between WSI pairs from different clusters. The proposed method introduces a two-stage contrastive learning approach WSI-level and patch-level contrastive learning to minimize these gaps effectively.
Comprehensive Pathological Image Segmentation via Teacher Aggregation for Tumor Microenvironment Analysis
Jan 06, 2025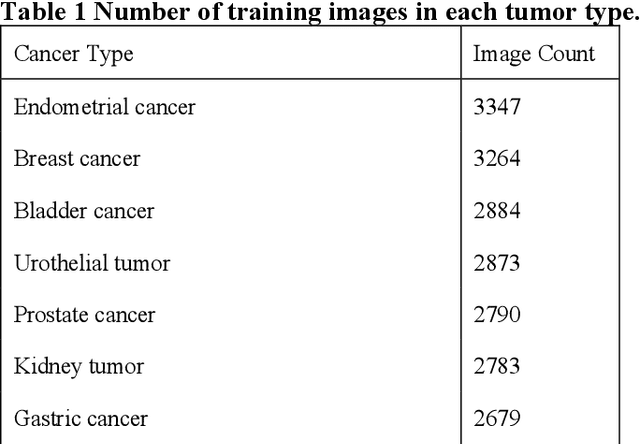


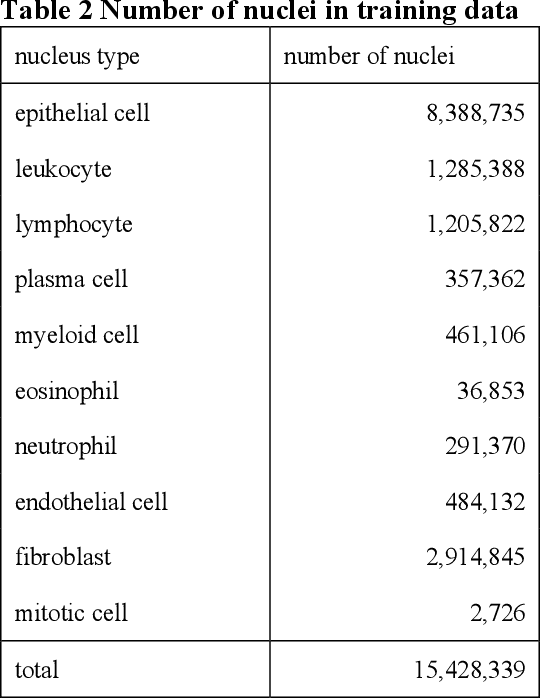
Abstract:The tumor microenvironment (TME) plays a crucial role in cancer progression and treatment response, yet current methods for its comprehensive analysis in H&E-stained tissue slides face significant limitations in the diversity of tissue cell types and accuracy. Here, we present PAGET (Pathological image segmentation via AGgrEgated Teachers), a new knowledge distillation approach that integrates multiple segmentation models while considering the hierarchical nature of cell types in the TME. By leveraging a unique dataset created through immunohistochemical restaining techniques and existing segmentation models, PAGET enables simultaneous identification and classification of 14 key TME components. We demonstrate PAGET's ability to perform rapid, comprehensive TME segmentation across various tissue types and medical institutions, advancing the quantitative analysis of tumor microenvironments. This method represents a significant step forward in enhancing our understanding of cancer biology and supporting precise clinical decision-making from large-scale histopathology images.
Cluster Entropy: Active Domain Adaptation in Pathological Image Segmentation
Apr 26, 2023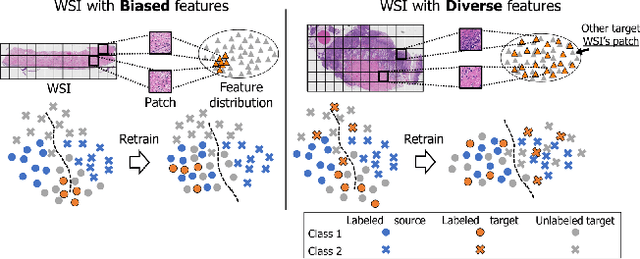
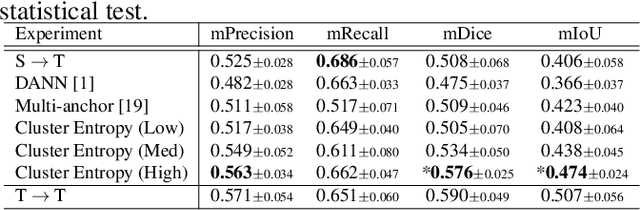

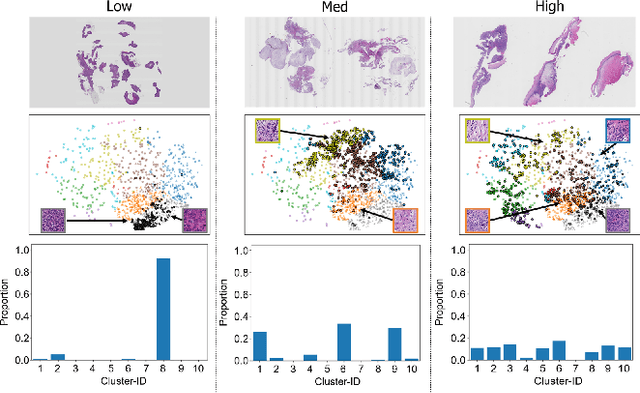
Abstract:The domain shift in pathological segmentation is an important problem, where a network trained by a source domain (collected at a specific hospital) does not work well in the target domain (from different hospitals) due to the different image features. Due to the problems of class imbalance and different class prior of pathology, typical unsupervised domain adaptation methods do not work well by aligning the distribution of source domain and target domain. In this paper, we propose a cluster entropy for selecting an effective whole slide image (WSI) that is used for semi-supervised domain adaptation. This approach can measure how the image features of the WSI cover the entire distribution of the target domain by calculating the entropy of each cluster and can significantly improve the performance of domain adaptation. Our approach achieved competitive results against the prior arts on datasets collected from two hospitals.
Domain Adaptive Multiple Instance Learning for Instance-level Prediction of Pathological Images
Apr 07, 2023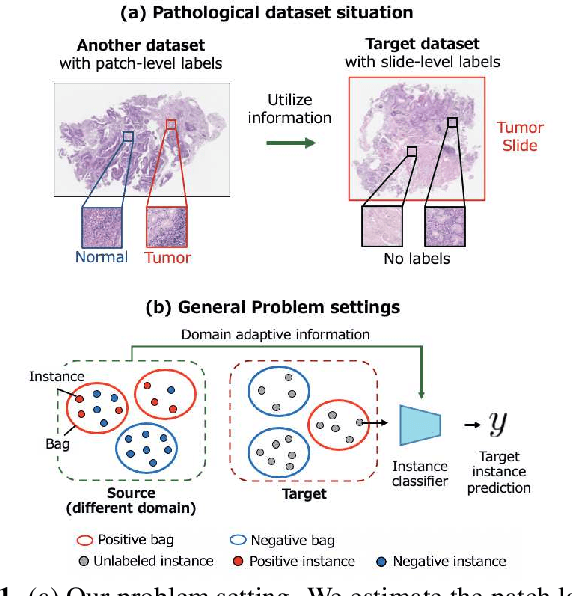
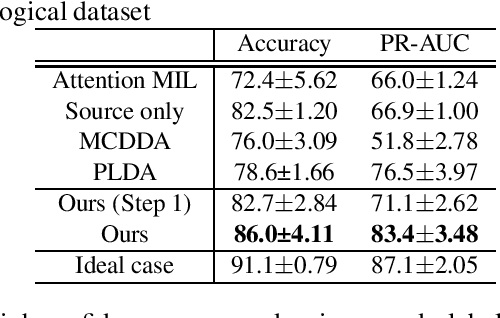
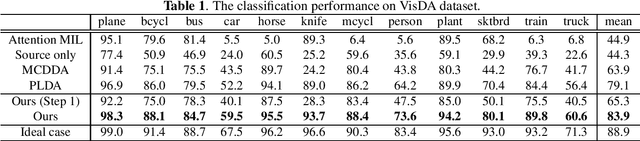
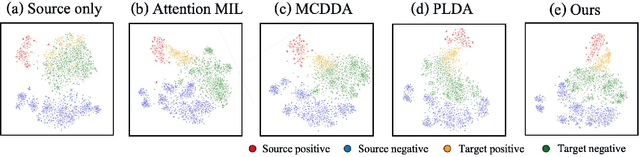
Abstract:Pathological image analysis is an important process for detecting abnormalities such as cancer from cell images. However, since the image size is generally very large, the cost of providing detailed annotations is high, which makes it difficult to apply machine learning techniques. One way to improve the performance of identifying abnormalities while keeping the annotation cost low is to use only labels for each slide, or to use information from another dataset that has already been labeled. However, such weak supervisory information often does not provide sufficient performance. In this paper, we proposed a new task setting to improve the classification performance of the target dataset without increasing annotation costs. And to solve this problem, we propose a pipeline that uses multiple instance learning (MIL) and domain adaptation (DA) methods. Furthermore, in order to combine the supervisory information of both methods effectively, we propose a method to create pseudo-labels with high confidence. We conducted experiments on the pathological image dataset we created for this study and showed that the proposed method significantly improves the classification performance compared to existing methods.
Cluster-Guided Semi-Supervised Domain Adaptation for Imbalanced Medical Image Classification
Mar 02, 2023Abstract:Semi-supervised domain adaptation is a technique to build a classifier for a target domain by modifying a classifier in another (source) domain using many unlabeled samples and a small number of labeled samples from the target domain. In this paper, we develop a semi-supervised domain adaptation method, which has robustness to class-imbalanced situations, which are common in medical image classification tasks. For robustness, we propose a weakly-supervised clustering pipeline to obtain high-purity clusters and utilize the clusters in representation learning for domain adaptation. The proposed method showed state-of-the-art performance in the experiment using severely class-imbalanced pathological image patches.
Achieving Transparency in Distributed Machine Learning with Explainable Data Collaboration
Dec 06, 2022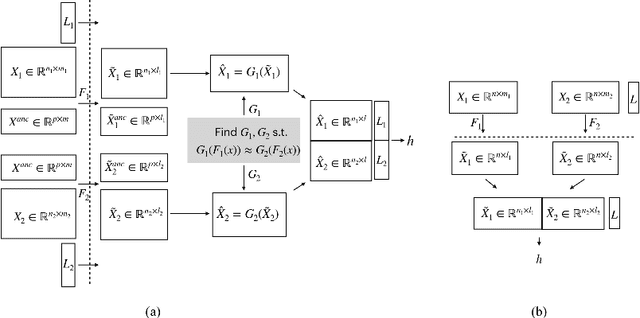
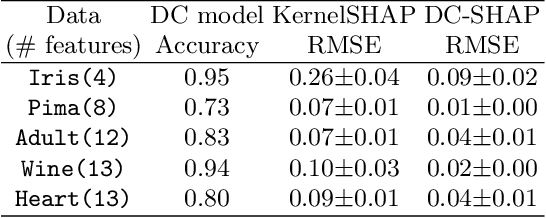
Abstract:Transparency of Machine Learning models used for decision support in various industries becomes essential for ensuring their ethical use. To that end, feature attribution methods such as SHAP (SHapley Additive exPlanations) are widely used to explain the predictions of black-box machine learning models to customers and developers. However, a parallel trend has been to train machine learning models in collaboration with other data holders without accessing their data. Such models, trained over horizontally or vertically partitioned data, present a challenge for explainable AI because the explaining party may have a biased view of background data or a partial view of the feature space. As a result, explanations obtained from different participants of distributed machine learning might not be consistent with one another, undermining trust in the product. This paper presents an Explainable Data Collaboration Framework based on a model-agnostic additive feature attribution algorithm (KernelSHAP) and Data Collaboration method of privacy-preserving distributed machine learning. In particular, we present three algorithms for different scenarios of explainability in Data Collaboration and verify their consistency with experiments on open-access datasets. Our results demonstrated a significant (by at least a factor of 1.75) decrease in feature attribution discrepancies among the users of distributed machine learning.
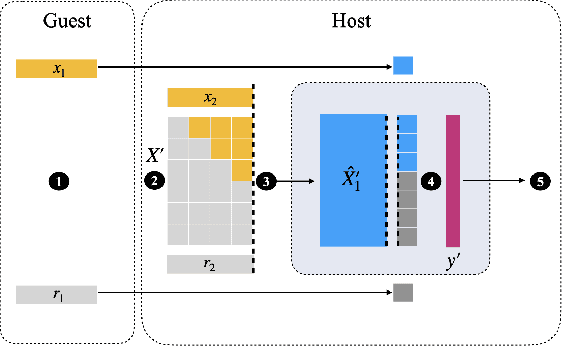
Non-readily identifiable data collaboration analysis for multiple datasets including personal information
Aug 31, 2022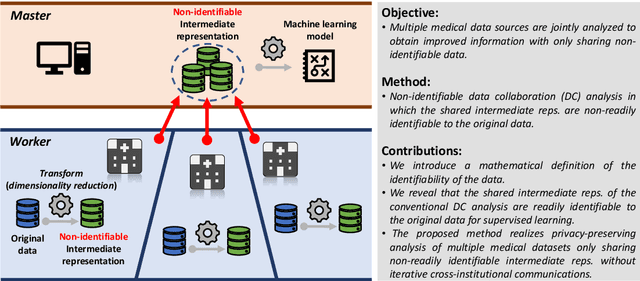
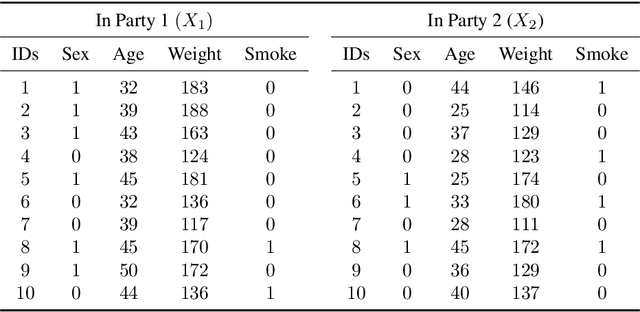
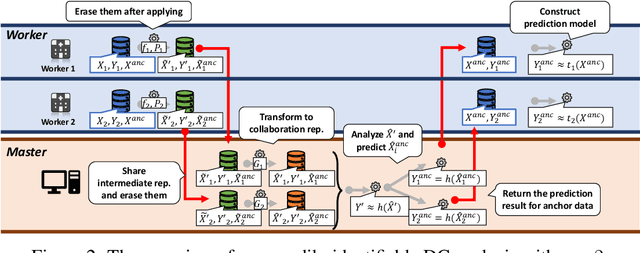
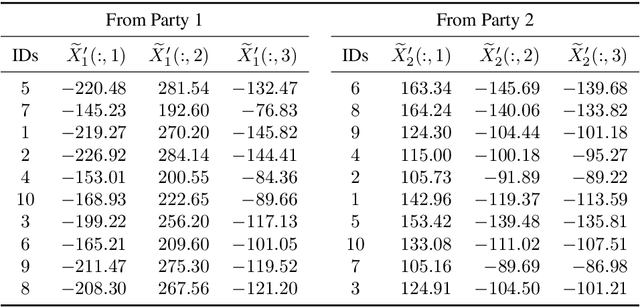
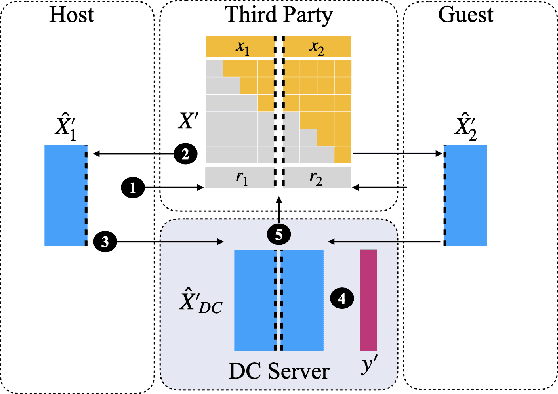
Abstract:Multi-source data fusion, in which multiple data sources are jointly analyzed to obtain improved information, has considerable research attention. For the datasets of multiple medical institutions, data confidentiality and cross-institutional communication are critical. In such cases, data collaboration (DC) analysis by sharing dimensionality-reduced intermediate representations without iterative cross-institutional communications may be appropriate. Identifiability of the shared data is essential when analyzing data including personal information. In this study, the identifiability of the DC analysis is investigated. The results reveals that the shared intermediate representations are readily identifiable to the original data for supervised learning. This study then proposes a non-readily identifiable DC analysis only sharing non-readily identifiable data for multiple medical datasets including personal information. The proposed method solves identifiability concerns based on a random sample permutation, the concept of interpretable DC analysis, and usage of functions that cannot be reconstructed. In numerical experiments on medical datasets, the proposed method exhibits a non-readily identifiability while maintaining a high recognition performance of the conventional DC analysis. For a hospital dataset, the proposed method exhibits a nine percentage point improvement regarding the recognition performance over the local analysis that uses only local dataset.
Multi-Stage Pathological Image Classification using Semantic Segmentation
Oct 10, 2019
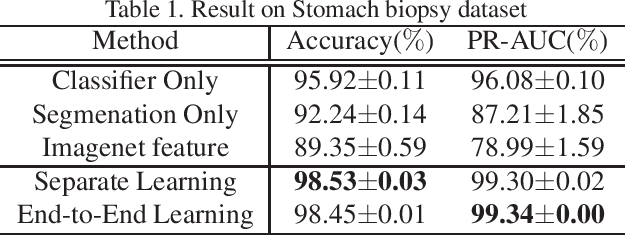
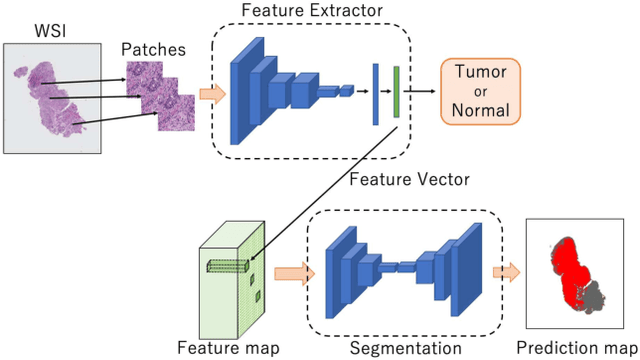
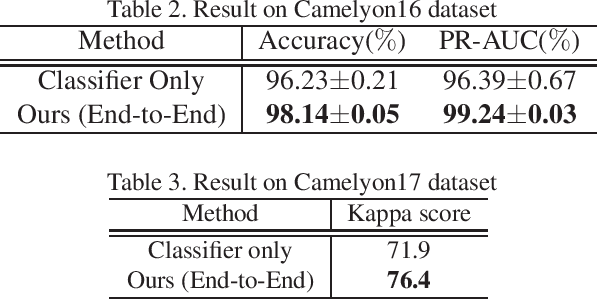
Abstract:Histopathological image analysis is an essential process for the discovery of diseases such as cancer. However, it is challenging to train CNN on whole slide images (WSIs) of gigapixel resolution considering the available memory capacity. Most of the previous works divide high resolution WSIs into small image patches and separately input them into the model to classify it as a tumor or a normal tissue. However, patch-based classification uses only patch-scale local information but ignores the relationship between neighboring patches. If we consider the relationship of neighboring patches and global features, we can improve the classification performance. In this paper, we propose a new model structure combining the patch-based classification model and whole slide-scale segmentation model in order to improve the prediction performance of automatic pathological diagnosis. We extract patch features from the classification model and input them into the segmentation model to obtain a whole slide tumor probability heatmap. The classification model considers patch-scale local features, and the segmentation model can take global information into account. We also propose a new optimization method that retains gradient information and trains the model partially for end-to-end learning with limited GPU memory capacity. We apply our method to the tumor/normal prediction on WSIs and the classification performance is improved compared with the conventional patch-based method.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge