Xiaoqing Liu
HE-SNR: Uncovering Latent Logic via Entropy for Guiding Mid-Training on SWE-BENCH
Jan 28, 2026Abstract:SWE-bench has emerged as the premier benchmark for evaluating Large Language Models on complex software engineering tasks. While these capabilities are fundamentally acquired during the mid-training phase and subsequently elicited during Supervised Fine-Tuning (SFT), there remains a critical deficit in metrics capable of guiding mid-training effectively. Standard metrics such as Perplexity (PPL) are compromised by the "Long-Context Tax" and exhibit weak correlation with downstream SWE performance. In this paper, we bridge this gap by first introducing a rigorous data filtering strategy. Crucially, we propose the Entropy Compression Hypothesis, redefining intelligence not by scalar Top-1 compression, but by the capacity to structure uncertainty into Entropy-Compressed States of low orders ("reasonable hesitation"). Grounded in this fine-grained entropy analysis, we formulate a novel metric, HE-SNR (High-Entropy Signal-to-Noise Ratio). Validated on industrial-scale Mixture-of-Experts (MoE) models across varying context windows (32K/128K), our approach demonstrates superior robustness and predictive power. This work provides both the theoretical foundation and practical tools for optimizing the latent potential of LLMs in complex engineering domains.
An Automatic Detection Method for Hematoma Features in Placental Abruption Ultrasound Images Based on Few-Shot Learning
Oct 24, 2025Abstract:Placental abruption is a severe complication during pregnancy, and its early accurate diagnosis is crucial for ensuring maternal and fetal safety. Traditional ultrasound diagnostic methods heavily rely on physician experience, leading to issues such as subjective bias and diagnostic inconsistencies. This paper proposes an improved model, EH-YOLOv11n (Enhanced Hemorrhage-YOLOv11n), based on small-sample learning, aiming to achieve automatic detection of hematoma features in placental ultrasound images. The model enhances performance through multidimensional optimization: it integrates wavelet convolution and coordinate convolution to strengthen frequency and spatial feature extraction; incorporates a cascaded group attention mechanism to suppress ultrasound artifacts and occlusion interference, thereby improving bounding box localization accuracy. Experimental results demonstrate a detection accuracy of 78%, representing a 2.5% improvement over YOLOv11n and a 13.7% increase over YOLOv8. The model exhibits significant superiority in precision-recall curves, confidence scores, and occlusion scenarios. Combining high accuracy with real-time processing, this model provides a reliable solution for computer-aided diagnosis of placental abruption, holding significant clinical application value.
RoHyDR: Robust Hybrid Diffusion Recovery for Incomplete Multimodal Emotion Recognition
May 23, 2025Abstract:Multimodal emotion recognition analyzes emotions by combining data from multiple sources. However, real-world noise or sensor failures often cause missing or corrupted data, creating the Incomplete Multimodal Emotion Recognition (IMER) challenge. In this paper, we propose Robust Hybrid Diffusion Recovery (RoHyDR), a novel framework that performs missing-modality recovery at unimodal, multimodal, feature, and semantic levels. For unimodal representation recovery of missing modalities, RoHyDR exploits a diffusion-based generator to generate distribution-consistent and semantically aligned representations from Gaussian noise, using available modalities as conditioning. For multimodal fusion recovery, we introduce adversarial learning to produce a realistic fused multimodal representation and recover missing semantic content. We further propose a multi-stage optimization strategy that enhances training stability and efficiency. In contrast to previous work, the hybrid diffusion and adversarial learning-based recovery mechanism in RoHyDR allows recovery of missing information in both unimodal representation and multimodal fusion, at both feature and semantic levels, effectively mitigating performance degradation caused by suboptimal optimization. Comprehensive experiments conducted on two widely used multimodal emotion recognition benchmarks demonstrate that our proposed method outperforms state-of-the-art IMER methods, achieving robust recognition performance under various missing-modality scenarios. Our code will be made publicly available upon acceptance.
SDR-Former: A Siamese Dual-Resolution Transformer for Liver Lesion Classification Using 3D Multi-Phase Imaging
Feb 27, 2024



Abstract:Automated classification of liver lesions in multi-phase CT and MR scans is of clinical significance but challenging. This study proposes a novel Siamese Dual-Resolution Transformer (SDR-Former) framework, specifically designed for liver lesion classification in 3D multi-phase CT and MR imaging with varying phase counts. The proposed SDR-Former utilizes a streamlined Siamese Neural Network (SNN) to process multi-phase imaging inputs, possessing robust feature representations while maintaining computational efficiency. The weight-sharing feature of the SNN is further enriched by a hybrid Dual-Resolution Transformer (DR-Former), comprising a 3D Convolutional Neural Network (CNN) and a tailored 3D Transformer for processing high- and low-resolution images, respectively. This hybrid sub-architecture excels in capturing detailed local features and understanding global contextual information, thereby, boosting the SNN's feature extraction capabilities. Additionally, a novel Adaptive Phase Selection Module (APSM) is introduced, promoting phase-specific intercommunication and dynamically adjusting each phase's influence on the diagnostic outcome. The proposed SDR-Former framework has been validated through comprehensive experiments on two clinical datasets: a three-phase CT dataset and an eight-phase MR dataset. The experimental results affirm the efficacy of the proposed framework. To support the scientific community, we are releasing our extensive multi-phase MR dataset for liver lesion analysis to the public. This pioneering dataset, being the first publicly available multi-phase MR dataset in this field, also underpins the MICCAI LLD-MMRI Challenge. The dataset is accessible at:https://bit.ly/3IyYlgN.
Generate to Understand for Representation
Jun 14, 2023Abstract:In recent years, a significant number of high-quality pretrained models have emerged, greatly impacting Natural Language Understanding (NLU), Natural Language Generation (NLG), and Text Representation tasks. Traditionally, these models are pretrained on custom domain corpora and finetuned for specific tasks, resulting in high costs related to GPU usage and labor. Unfortunately, recent trends in language modeling have shifted towards enhancing performance through scaling, further exacerbating the associated costs. Introducing GUR: a pretraining framework that combines language modeling and contrastive learning objectives in a single training step. We select similar text pairs based on their Longest Common Substring (LCS) from raw unlabeled documents and train the model using masked language modeling and unsupervised contrastive learning. The resulting model, GUR, achieves impressive results without any labeled training data, outperforming all other pretrained baselines as a retriever at the recall benchmark in a zero-shot setting. Additionally, GUR maintains its language modeling ability, as demonstrated in our ablation experiment. Our code is available at \url{https://github.com/laohur/GUR}.
Mixing Data Augmentation with Preserving Foreground Regions in Medical Image Segmentation
Apr 26, 2023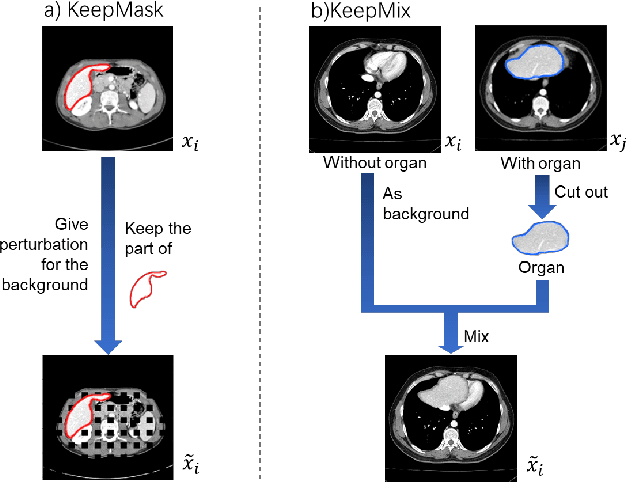
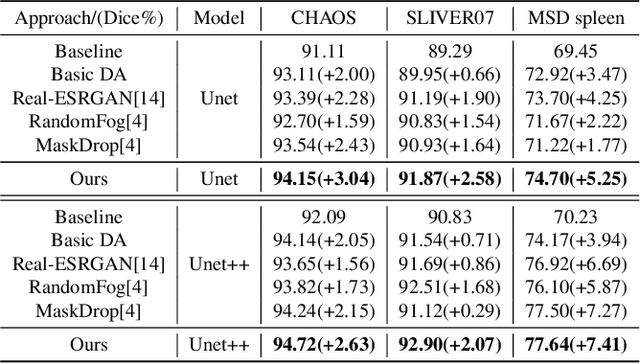
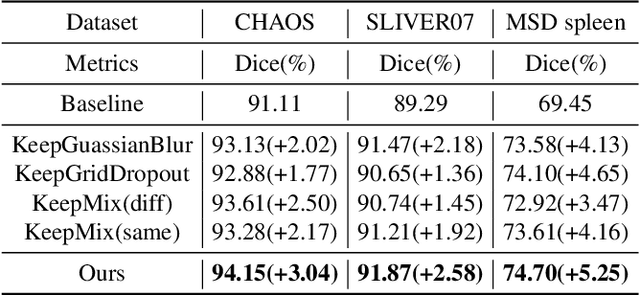
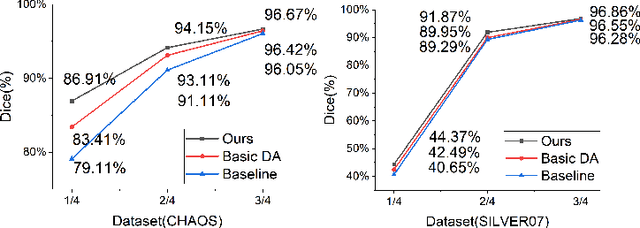
Abstract:The development of medical image segmentation using deep learning can significantly support doctors' diagnoses. Deep learning needs large amounts of data for training, which also requires data augmentation to extend diversity for preventing overfitting. However, the existing methods for data augmentation of medical image segmentation are mainly based on models which need to update parameters and cost extra computing resources. We proposed data augmentation methods designed to train a high accuracy deep learning network for medical image segmentation. The proposed data augmentation approaches are called KeepMask and KeepMix, which can create medical images by better identifying the boundary of the organ with no more parameters. Our methods achieved better performance and obtained more precise boundaries for medical image segmentation on datasets. The dice coefficient of our methods achieved 94.15% (3.04% higher than baseline) on CHAOS and 74.70% (5.25% higher than baseline) on MSD spleen with Unet.
Cluster Entropy: Active Domain Adaptation in Pathological Image Segmentation
Apr 26, 2023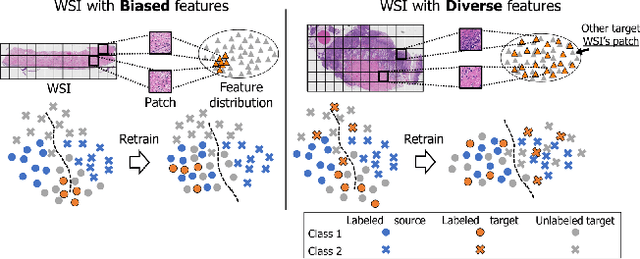
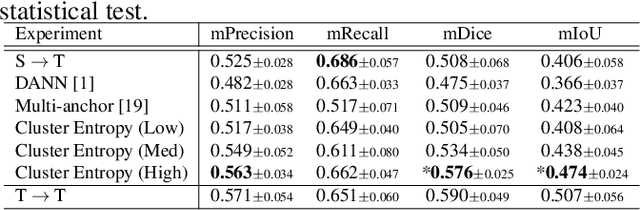

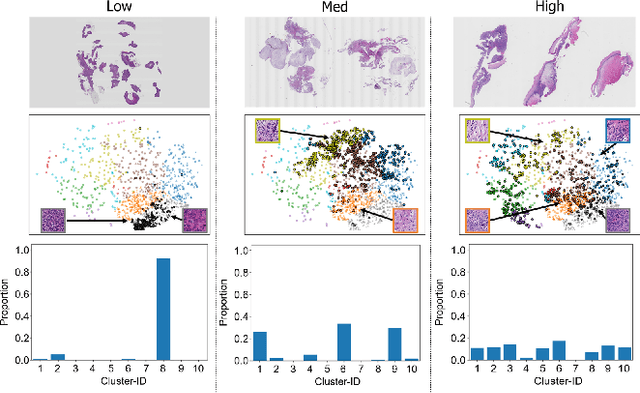
Abstract:The domain shift in pathological segmentation is an important problem, where a network trained by a source domain (collected at a specific hospital) does not work well in the target domain (from different hospitals) due to the different image features. Due to the problems of class imbalance and different class prior of pathology, typical unsupervised domain adaptation methods do not work well by aligning the distribution of source domain and target domain. In this paper, we propose a cluster entropy for selecting an effective whole slide image (WSI) that is used for semi-supervised domain adaptation. This approach can measure how the image features of the WSI cover the entire distribution of the target domain by calculating the entropy of each cluster and can significantly improve the performance of domain adaptation. Our approach achieved competitive results against the prior arts on datasets collected from two hospitals.
Compound Domain Generalization via Meta-Knowledge Encoding
Mar 24, 2022



Abstract:Domain generalization (DG) aims to improve the generalization performance for an unseen target domain by using the knowledge of multiple seen source domains. Mainstream DG methods typically assume that the domain label of each source sample is known a priori, which is challenged to be satisfied in many real-world applications. In this paper, we study a practical problem of compound DG, which relaxes the discrete domain assumption to the mixed source domains setting. On the other hand, current DG algorithms prioritize the focus on semantic invariance across domains (one-vs-one), while paying less attention to the holistic semantic structure (many-vs-many). Such holistic semantic structure, referred to as meta-knowledge here, is crucial for learning generalizable representations. To this end, we present Compound Domain Generalization via Meta-Knowledge Encoding (COMEN), a general approach to automatically discover and model latent domains in two steps. Firstly, we introduce Style-induced Domain-specific Normalization (SDNorm) to re-normalize the multi-modal underlying distributions, thereby dividing the mixture of source domains into latent clusters. Secondly, we harness the prototype representations, the centroids of classes, to perform relational modeling in the embedding space with two parallel and complementary modules, which explicitly encode the semantic structure for the out-of-distribution generalization. Experiments on four standard DG benchmarks reveal that COMEN exceeds the state-of-the-art performance without the need of domain supervision.
Identification of Pediatric Respiratory Diseases Using Fine-grained Diagnosis System
Aug 24, 2021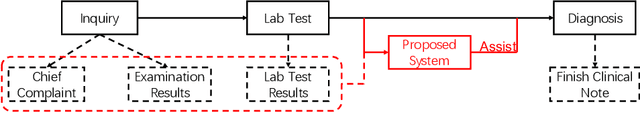
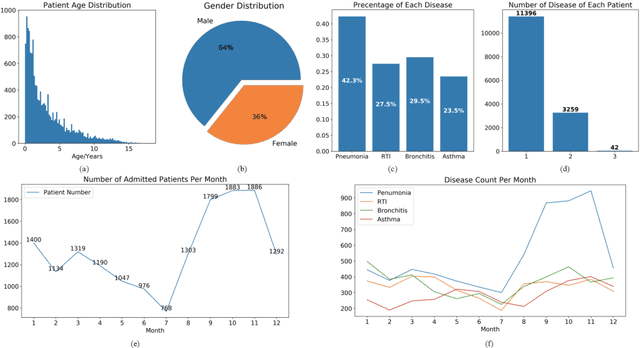
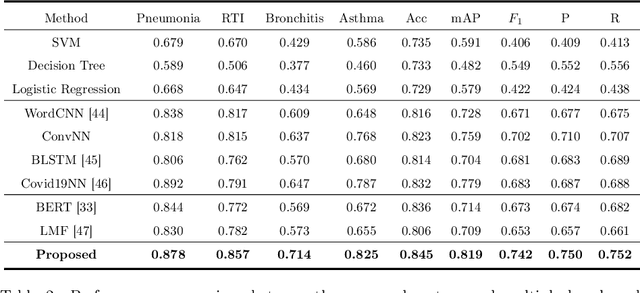
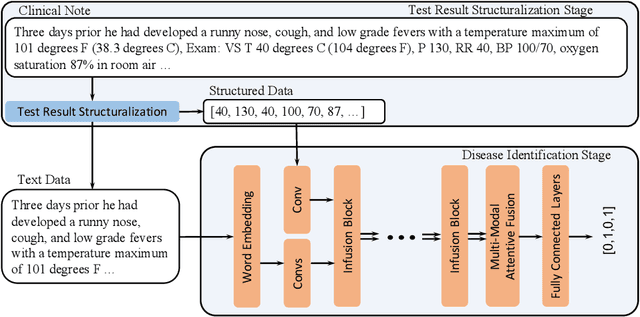
Abstract:Respiratory diseases, including asthma, bronchitis, pneumonia, and upper respiratory tract infection (RTI), are among the most common diseases in clinics. The similarities among the symptoms of these diseases precludes prompt diagnosis upon the patients' arrival. In pediatrics, the patients' limited ability in expressing their situation makes precise diagnosis even harder. This becomes worse in primary hospitals, where the lack of medical imaging devices and the doctors' limited experience further increase the difficulty of distinguishing among similar diseases. In this paper, a pediatric fine-grained diagnosis-assistant system is proposed to provide prompt and precise diagnosis using solely clinical notes upon admission, which would assist clinicians without changing the diagnostic process. The proposed system consists of two stages: a test result structuralization stage and a disease identification stage. The first stage structuralizes test results by extracting relevant numerical values from clinical notes, and the disease identification stage provides a diagnosis based on text-form clinical notes and the structured data obtained from the first stage. A novel deep learning algorithm was developed for the disease identification stage, where techniques including adaptive feature infusion and multi-modal attentive fusion were introduced to fuse structured and text data together. Clinical notes from over 12000 patients with respiratory diseases were used to train a deep learning model, and clinical notes from a non-overlapping set of about 1800 patients were used to evaluate the performance of the trained model. The average precisions (AP) for pneumonia, RTI, bronchitis and asthma are 0.878, 0.857, 0.714, and 0.825, respectively, achieving a mean AP (mAP) of 0.819.
A Structure-Aware Relation Network for Thoracic Diseases Detection and Segmentation
Apr 21, 2021



Abstract:Instance level detection and segmentation of thoracic diseases or abnormalities are crucial for automatic diagnosis in chest X-ray images. Leveraging on constant structure and disease relations extracted from domain knowledge, we propose a structure-aware relation network (SAR-Net) extending Mask R-CNN. The SAR-Net consists of three relation modules: 1. the anatomical structure relation module encoding spatial relations between diseases and anatomical parts. 2. the contextual relation module aggregating clues based on query-key pair of disease RoI and lung fields. 3. the disease relation module propagating co-occurrence and causal relations into disease proposals. Towards making a practical system, we also provide ChestX-Det, a chest X-Ray dataset with instance-level annotations (boxes and masks). ChestX-Det is a subset of the public dataset NIH ChestX-ray14. It contains ~3500 images of 13 common disease categories labeled by three board-certified radiologists. We evaluate our SAR-Net on it and another dataset DR-Private. Experimental results show that it can enhance the strong baseline of Mask R-CNN with significant improvements. The ChestX-Det is released at https://github.com/Deepwise-AILab/ChestX-Det-Dataset.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge