Ryoma Bise
Hypernetwork-Based Adaptive Aggregation for Multimodal Multiple-Instance Learning in Predicting Coronary Calcium Debulking
Jan 29, 2026Abstract:In this paper, we present the first attempt to estimate the necessity of debulking coronary artery calcifications from computed tomography (CT) images. We formulate this task as a Multiple-instance Learning (MIL) problem. The difficulty of this task lies in that physicians adjust their focus and decision criteria for device usage according to tabular data representing each patient's condition. To address this issue, we propose a hypernetwork-based adaptive aggregation transformer (HyperAdAgFormer), which adaptively modifies the feature aggregation strategy for each patient based on tabular data through a hypernetwork. The experiments using the clinical dataset demonstrated the effectiveness of HyperAdAgFormer. The code is publicly available at https://github.com/Shiku-Kaito/HyperAdAgFormer.
Domain Adaptation for Ulcerative Colitis Severity Estimation Using Patient-Level Diagnoses
Sep 18, 2025



Abstract:The development of methods to estimate the severity of Ulcerative Colitis (UC) is of significant importance. However, these methods often suffer from domain shifts caused by differences in imaging devices and clinical settings across hospitals. Although several domain adaptation methods have been proposed to address domain shift, they still struggle with the lack of supervision in the target domain or the high cost of annotation. To overcome these challenges, we propose a novel Weakly Supervised Domain Adaptation method that leverages patient-level diagnostic results, which are routinely recorded in UC diagnosis, as weak supervision in the target domain. The proposed method aligns class-wise distributions across domains using Shared Aggregation Tokens and a Max-Severity Triplet Loss, which leverages the characteristic that patient-level diagnoses are determined by the most severe region within each patient. Experimental results demonstrate that our method outperforms comparative DA approaches, improving UC severity estimation in a domain-shifted setting.
Learning from Majority Label: A Novel Problem in Multi-class Multiple-Instance Learning
Sep 04, 2025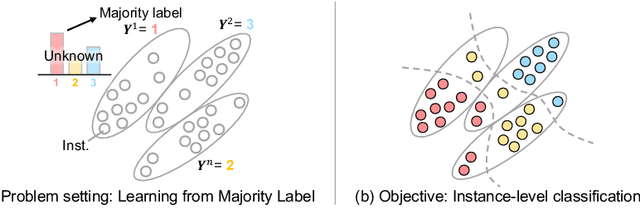

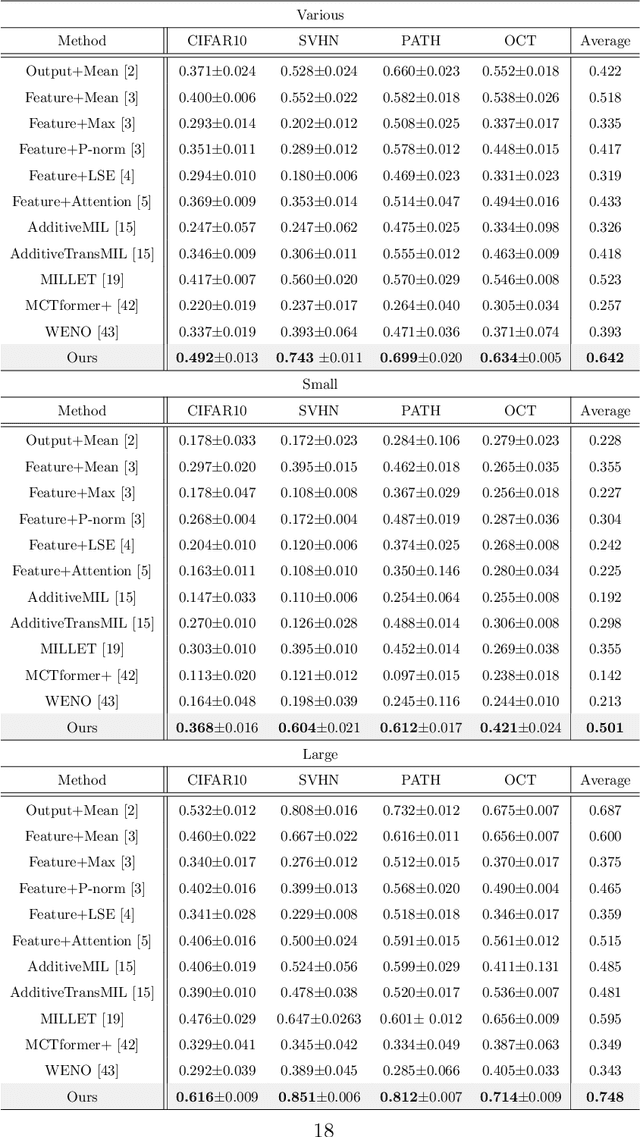
Abstract:The paper proposes a novel multi-class Multiple-Instance Learning (MIL) problem called Learning from Majority Label (LML). In LML, the majority class of instances in a bag is assigned as the bag-level label. The goal of LML is to train a classification model that estimates the class of each instance using the majority label. This problem is valuable in a variety of applications, including pathology image segmentation, political voting prediction, customer sentiment analysis, and environmental monitoring. To solve LML, we propose a Counting Network trained to produce bag-level majority labels, estimated by counting the number of instances in each class. Furthermore, analysis experiments on the characteristics of LML revealed that bags with a high proportion of the majority class facilitate learning. Based on this result, we developed a Majority Proportion Enhancement Module (MPEM) that increases the proportion of the majority class by removing minority class instances within the bags. Experiments demonstrate the superiority of the proposed method on four datasets compared to conventional MIL methods. Moreover, ablation studies confirmed the effectiveness of each module. The code is available at \href{https://github.com/Shiku-Kaito/Learning-from-Majority-Label-A-Novel-Problem-in-Multi-class-Multiple-Instance-Learning}{here}.
Adaptive Pseudo Label Selection for Individual Unlabeled Data by Positive and Unlabeled Learning
Aug 11, 2025Abstract:This paper proposes a novel pseudo-labeling method for medical image segmentation that can perform learning on ``individual images'' to select effective pseudo-labels. We introduce Positive and Unlabeled Learning (PU learning), which uses only positive and unlabeled data for binary classification problems, to obtain the appropriate metric for discriminating foreground and background regions on each unlabeled image. Our PU learning makes us easy to select pseudo-labels for various background regions. The experimental results show the effectiveness of our method.
Domain Generalization of Pathological Image Segmentation by Patch-Level and WSI-Level Contrastive Learning
Aug 11, 2025


Abstract:In this paper, we address domain shifts in pathological images by focusing on shifts within whole slide images~(WSIs), such as patient characteristics and tissue thickness, rather than shifts between hospitals. Traditional approaches rely on multi-hospital data, but data collection challenges often make this impractical. Therefore, the proposed domain generalization method captures and leverages intra-hospital domain shifts by clustering WSI-level features from non-tumor regions and treating these clusters as domains. To mitigate domain shift, we apply contrastive learning to reduce feature gaps between WSI pairs from different clusters. The proposed method introduces a two-stage contrastive learning approach WSI-level and patch-level contrastive learning to minimize these gaps effectively.
Enhancing Reliability of Medical Image Diagnosis through Top-rank Learning with Rejection Module
Aug 11, 2025Abstract:In medical image processing, accurate diagnosis is of paramount importance. Leveraging machine learning techniques, particularly top-rank learning, shows significant promise by focusing on the most crucial instances. However, challenges arise from noisy labels and class-ambiguous instances, which can severely hinder the top-rank objective, as they may be erroneously placed among the top-ranked instances. To address these, we propose a novel approach that enhances toprank learning by integrating a rejection module. Cooptimized with the top-rank loss, this module identifies and mitigates the impact of outliers that hinder training effectiveness. The rejection module functions as an additional branch, assessing instances based on a rejection function that measures their deviation from the norm. Through experimental validation on a medical dataset, our methodology demonstrates its efficacy in detecting and mitigating outliers, improving the reliability and accuracy of medical image diagnoses.
Towards Spatial Transcriptomics-guided Pathological Image Recognition with Batch-Agnostic Encoder
Mar 10, 2025



Abstract:Spatial transcriptomics (ST) is a novel technique that simultaneously captures pathological images and gene expression profiling with spatial coordinates. Since ST is closely related to pathological features such as disease subtypes, it may be valuable to augment image representation with pathological information. However, there are no attempts to leverage ST for image recognition ({\it i.e,} patch-level classification of subtypes of pathological image.). One of the big challenges is significant batch effects in spatial transcriptomics that make it difficult to extract pathological features of images from ST. In this paper, we propose a batch-agnostic contrastive learning framework that can extract consistent signals from gene expression of ST in multiple patients. To extract consistent signals from ST, we utilize the batch-agnostic gene encoder that is trained in a variational inference manner. Experiments demonstrated the effectiveness of our framework on a publicly available dataset. Code is publicly available at https://github.com/naivete5656/TPIRBAE
Instance-wise Supervision-level Optimization in Active Learning
Mar 09, 2025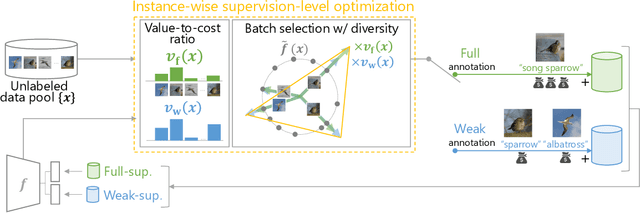
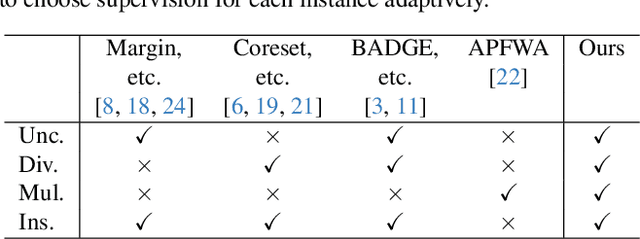
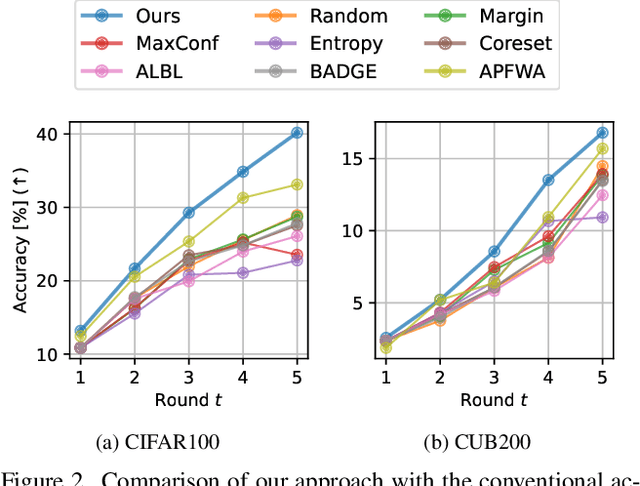
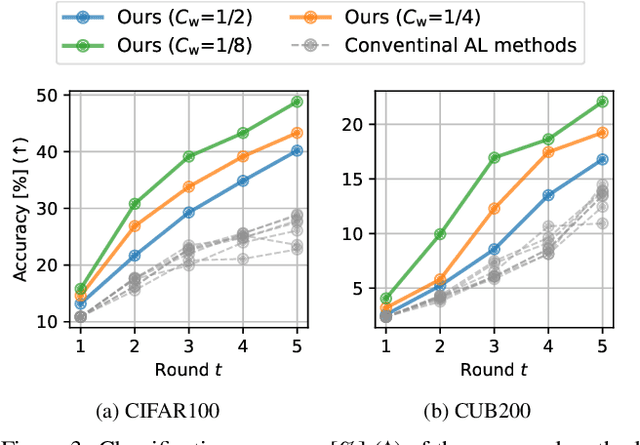
Abstract:Active learning (AL) is a label-efficient machine learning paradigm that focuses on selectively annotating high-value instances to maximize learning efficiency. Its effectiveness can be further enhanced by incorporating weak supervision, which uses rough yet cost-effective annotations instead of exact (i.e., full) but expensive annotations. We introduce a novel AL framework, Instance-wise Supervision-Level Optimization (ISO), which not only selects the instances to annotate but also determines their optimal annotation level within a fixed annotation budget. Its optimization criterion leverages the value-to-cost ratio (VCR) of each instance while ensuring diversity among the selected instances. In classification experiments, ISO consistently outperforms traditional AL methods and surpasses a state-of-the-art AL approach that combines full and weak supervision, achieving higher accuracy at a lower overall cost. This code is available at https://github.com/matsuo-shinnosuke/ISOAL.
Guidance-base Diffusion Models for Improving Photoacoustic Image Quality
Feb 10, 2025



Abstract:Photoacoustic(PA) imaging is a non-destructive and non-invasive technology for visualizing minute blood vessel structures in the body using ultrasonic sensors. In PA imaging, the image quality of a single-shot image is poor, and it is necessary to improve the image quality by averaging many single-shot images. Therefore, imaging the entire subject requires high imaging costs. In our study, we propose a method to improve the quality of PA images using diffusion models. In our method, we improve the reverse diffusion process using sensor information of PA imaging and introduce a guidance method using imaging condition information to generate high-quality images.
Ordinal Multiple-instance Learning for Ulcerative Colitis Severity Estimation with Selective Aggregated Transformer
Nov 22, 2024



Abstract:Patient-level diagnosis of severity in ulcerative colitis (UC) is common in real clinical settings, where the most severe score in a patient is recorded. However, previous UC classification methods (i.e., image-level estimation) mainly assumed the input was a single image. Thus, these methods can not utilize severity labels recorded in real clinical settings. In this paper, we propose a patient-level severity estimation method by a transformer with selective aggregator tokens, where a severity label is estimated from multiple images taken from a patient, similar to a clinical setting. Our method can effectively aggregate features of severe parts from a set of images captured in each patient, and it facilitates improving the discriminative ability between adjacent severity classes. Experiments demonstrate the effectiveness of the proposed method on two datasets compared with the state-of-the-art MIL methods. Moreover, we evaluated our method in real clinical settings and confirmed that our method outperformed the previous image-level methods. The code is publicly available at https://github.com/Shiku-Kaito/Ordinal-Multiple-instance-Learning-for-Ulcerative-Colitis-Severity-Estimation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge