Akihiko Yoshizawa
Domain Generalization of Pathological Image Segmentation by Patch-Level and WSI-Level Contrastive Learning
Aug 11, 2025


Abstract:In this paper, we address domain shifts in pathological images by focusing on shifts within whole slide images~(WSIs), such as patient characteristics and tissue thickness, rather than shifts between hospitals. Traditional approaches rely on multi-hospital data, but data collection challenges often make this impractical. Therefore, the proposed domain generalization method captures and leverages intra-hospital domain shifts by clustering WSI-level features from non-tumor regions and treating these clusters as domains. To mitigate domain shift, we apply contrastive learning to reduce feature gaps between WSI pairs from different clusters. The proposed method introduces a two-stage contrastive learning approach WSI-level and patch-level contrastive learning to minimize these gaps effectively.
Theoretical Proportion Label Perturbation for Learning from Label Proportions in Large Bags
Aug 26, 2024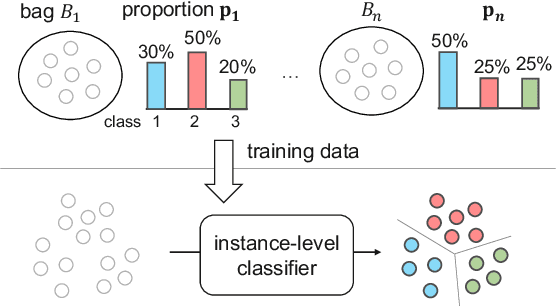
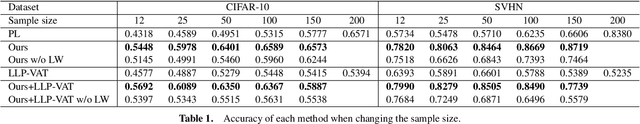
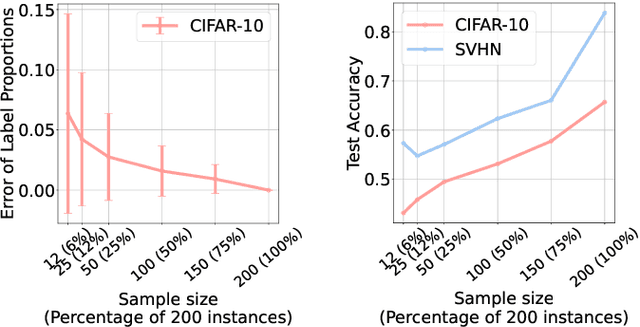
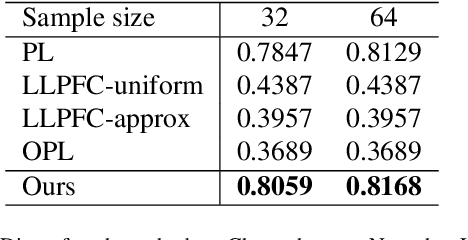
Abstract:Learning from label proportions (LLP) is a kind of weakly supervised learning that trains an instance-level classifier from label proportions of bags, which consist of sets of instances without using instance labels. A challenge in LLP arises when the number of instances in a bag (bag size) is numerous, making the traditional LLP methods difficult due to GPU memory limitations. This study aims to develop an LLP method capable of learning from bags with large sizes. In our method, smaller bags (mini-bags) are generated by sampling instances from large-sized bags (original bags), and these mini-bags are used in place of the original bags. However, the proportion of a mini-bag is unknown and differs from that of the original bag, leading to overfitting. To address this issue, we propose a perturbation method for the proportion labels of sampled mini-bags to mitigate overfitting to noisy label proportions. This perturbation is added based on the multivariate hypergeometric distribution, which is statistically modeled. Additionally, loss weighting is implemented to reduce the negative impact of proportions sampled from the tail of the distribution. Experimental results demonstrate that the proportion label perturbation and loss weighting achieve classification accuracy comparable to that obtained without sampling. Our codes are available at https://github.com/stainlessnight/LLP-LargeBags.
Learning from Partial Label Proportions for Whole Slide Image Segmentation
May 15, 2024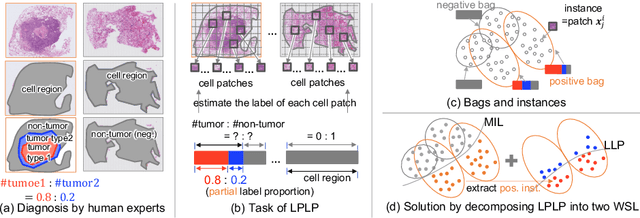

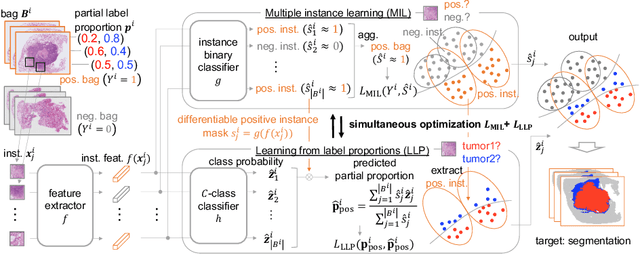
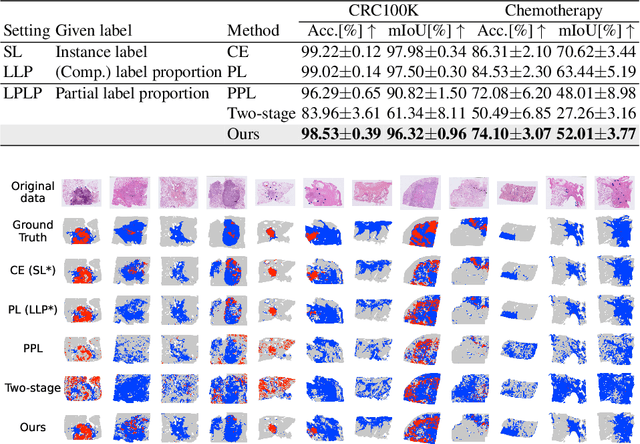
Abstract:In this paper, we address the segmentation of tumor subtypes in whole slide images (WSI) by utilizing incomplete label proportions. Specifically, we utilize `partial' label proportions, which give the proportions among tumor subtypes but do not give the proportion between tumor and non-tumor. Partial label proportions are recorded as the standard diagnostic information by pathologists, and we, therefore, want to use them for realizing the segmentation model that can classify each WSI patch into one of the tumor subtypes or non-tumor. We call this problem ``learning from partial label proportions (LPLP)'' and formulate the problem as a weakly supervised learning problem. Then, we propose an efficient algorithm for this challenging problem by decomposing it into two weakly supervised learning subproblems: multiple instance learning (MIL) and learning from label proportions (LLP). These subproblems are optimized efficiently in the end-to-end manner. The effectiveness of our algorithm is demonstrated through experiments conducted on two WSI datasets.
Proportion Estimation by Masked Learning from Label Proportion
May 08, 2024Abstract:The PD-L1 rate, the number of PD-L1 positive tumor cells over the total number of all tumor cells, is an important metric for immunotherapy. This metric is recorded as diagnostic information with pathological images. In this paper, we propose a proportion estimation method with a small amount of cell-level annotation and proportion annotation, which can be easily collected. Since the PD-L1 rate is calculated from only `tumor cells' and not using `non-tumor cells', we first detect tumor cells with a detection model. Then, we estimate the PD-L1 proportion by introducing a masking technique to `learning from label proportion.' In addition, we propose a weighted focal proportion loss to address data imbalance problems. Experiments using clinical data demonstrate the effectiveness of our method. Our method achieved the best performance in comparisons.
Cluster Entropy: Active Domain Adaptation in Pathological Image Segmentation
Apr 26, 2023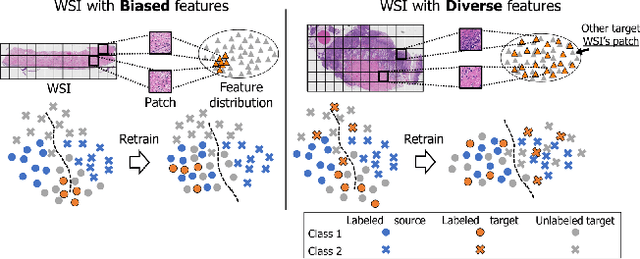
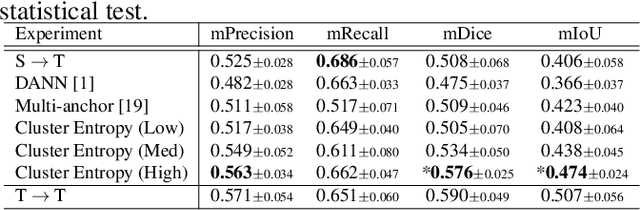

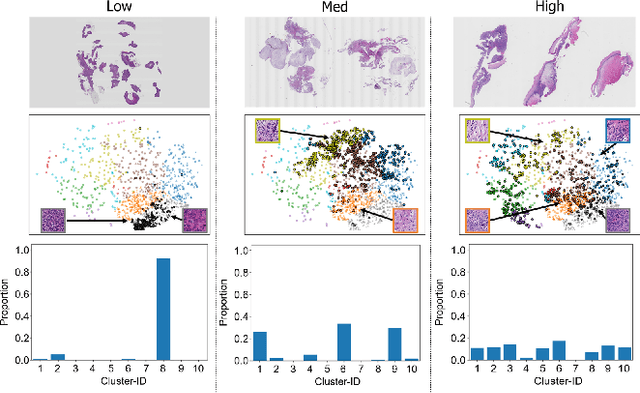
Abstract:The domain shift in pathological segmentation is an important problem, where a network trained by a source domain (collected at a specific hospital) does not work well in the target domain (from different hospitals) due to the different image features. Due to the problems of class imbalance and different class prior of pathology, typical unsupervised domain adaptation methods do not work well by aligning the distribution of source domain and target domain. In this paper, we propose a cluster entropy for selecting an effective whole slide image (WSI) that is used for semi-supervised domain adaptation. This approach can measure how the image features of the WSI cover the entire distribution of the target domain by calculating the entropy of each cluster and can significantly improve the performance of domain adaptation. Our approach achieved competitive results against the prior arts on datasets collected from two hospitals.
Domain Adaptive Multiple Instance Learning for Instance-level Prediction of Pathological Images
Apr 07, 2023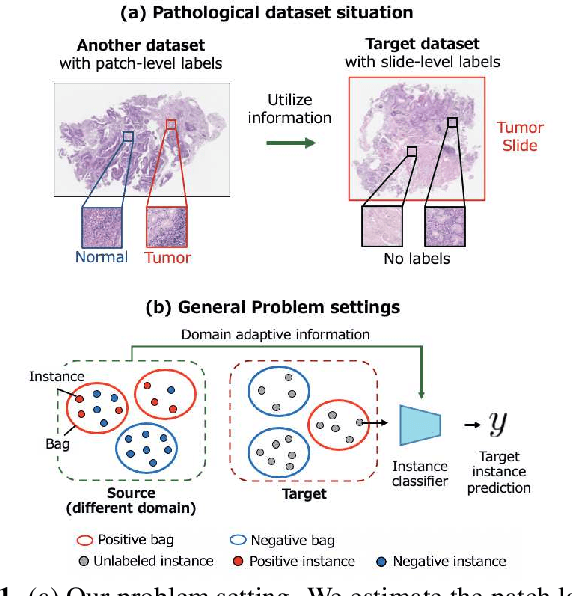
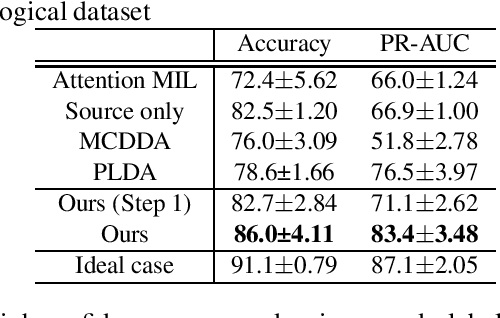
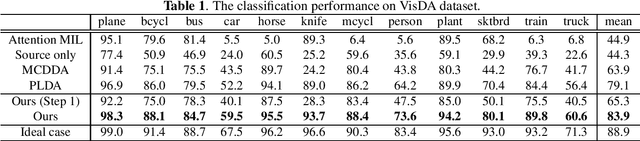
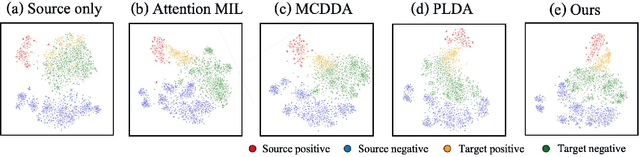
Abstract:Pathological image analysis is an important process for detecting abnormalities such as cancer from cell images. However, since the image size is generally very large, the cost of providing detailed annotations is high, which makes it difficult to apply machine learning techniques. One way to improve the performance of identifying abnormalities while keeping the annotation cost low is to use only labels for each slide, or to use information from another dataset that has already been labeled. However, such weak supervisory information often does not provide sufficient performance. In this paper, we proposed a new task setting to improve the classification performance of the target dataset without increasing annotation costs. And to solve this problem, we propose a pipeline that uses multiple instance learning (MIL) and domain adaptation (DA) methods. Furthermore, in order to combine the supervisory information of both methods effectively, we propose a method to create pseudo-labels with high confidence. We conducted experiments on the pathological image dataset we created for this study and showed that the proposed method significantly improves the classification performance compared to existing methods.
Cluster-Guided Semi-Supervised Domain Adaptation for Imbalanced Medical Image Classification
Mar 02, 2023Abstract:Semi-supervised domain adaptation is a technique to build a classifier for a target domain by modifying a classifier in another (source) domain using many unlabeled samples and a small number of labeled samples from the target domain. In this paper, we develop a semi-supervised domain adaptation method, which has robustness to class-imbalanced situations, which are common in medical image classification tasks. For robustness, we propose a weakly-supervised clustering pipeline to obtain high-purity clusters and utilize the clusters in representation learning for domain adaptation. The proposed method showed state-of-the-art performance in the experiment using severely class-imbalanced pathological image patches.
Patch-Based Cervical Cancer Segmentation using Distance from Boundary of Tissue
Aug 19, 2021

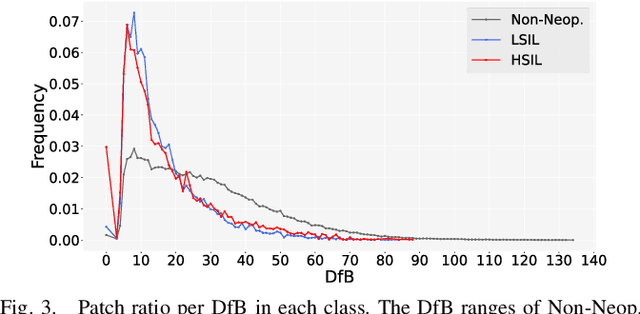

Abstract:Pathological diagnosis is used for examining cancer in detail, and its automation is in demand. To automatically segment each cancer area, a patch-based approach is usually used since a Whole Slide Image (WSI) is huge. However, this approach loses the global information needed to distinguish between classes. In this paper, we utilized the Distance from the Boundary of tissue (DfB), which is global information that can be extracted from the original image. We experimentally applied our method to the three-class classification of cervical cancer, and found that it improved the total performance compared with the conventional method.
Negative Pseudo Labeling using Class Proportion for Semantic Segmentation in Pathology
Jul 16, 2020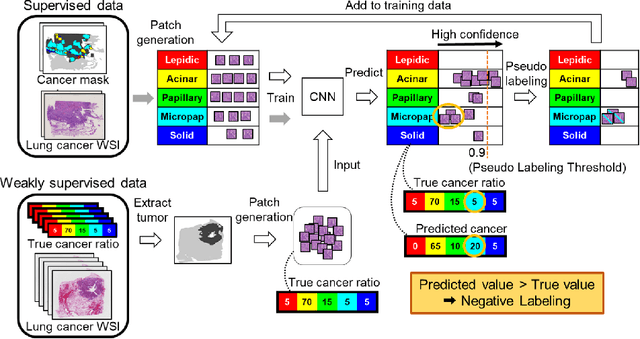
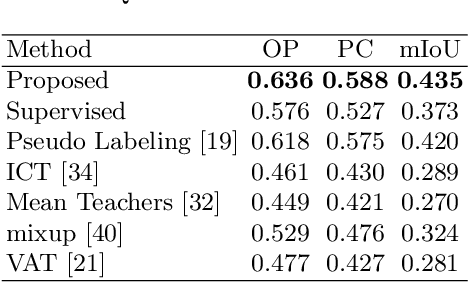
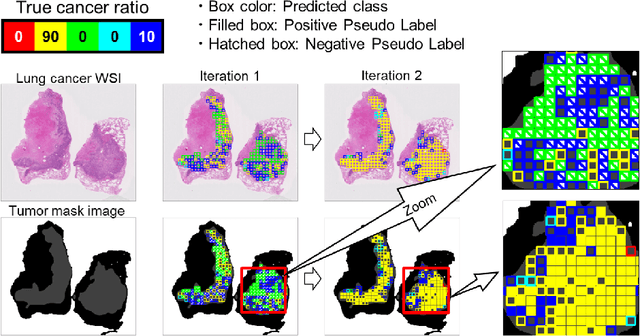
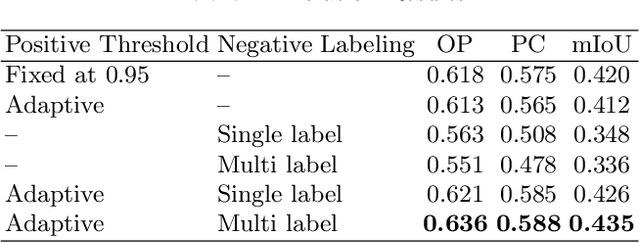
Abstract:We propose a weakly-supervised cell tracking method that can train a convolutional neural network (CNN) by using only the annotation of "cell detection" (i.e., the coordinates of cell positions) without association information, in which cell positions can be easily obtained by nuclear staining. First, we train a co-detection CNN that detects cells in successive frames by using weak-labels. Our key assumption is that the co-detection CNN implicitly learns association in addition to detection. To obtain the association information, we propose a backward-and-forward propagation method that analyzes the correspondence of cell positions in the detection maps output of the co-detection CNN. Experiments demonstrated that the proposed method can match positions by analyzing the co-detection CNN. Even though the method uses only weak supervision, the performance of our method was almost the same as the state-of-the-art supervised method.
Multi-Stage Pathological Image Classification using Semantic Segmentation
Oct 10, 2019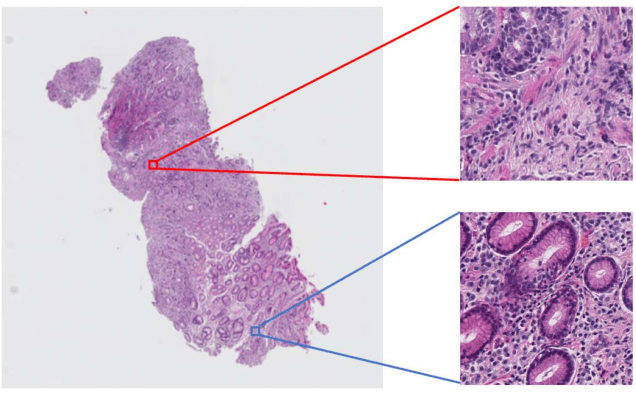
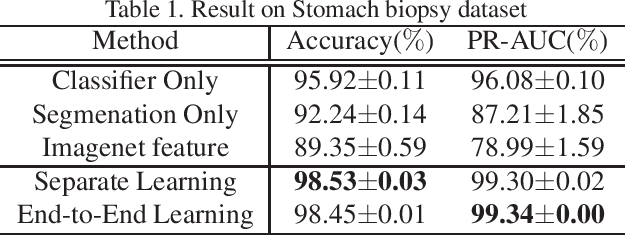
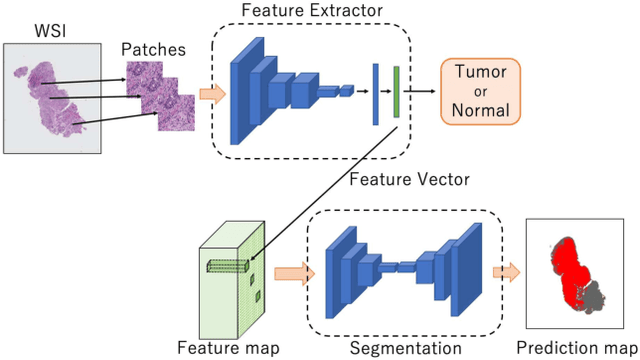
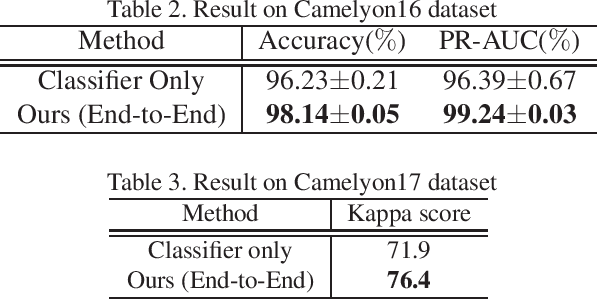
Abstract:Histopathological image analysis is an essential process for the discovery of diseases such as cancer. However, it is challenging to train CNN on whole slide images (WSIs) of gigapixel resolution considering the available memory capacity. Most of the previous works divide high resolution WSIs into small image patches and separately input them into the model to classify it as a tumor or a normal tissue. However, patch-based classification uses only patch-scale local information but ignores the relationship between neighboring patches. If we consider the relationship of neighboring patches and global features, we can improve the classification performance. In this paper, we propose a new model structure combining the patch-based classification model and whole slide-scale segmentation model in order to improve the prediction performance of automatic pathological diagnosis. We extract patch features from the classification model and input them into the segmentation model to obtain a whole slide tumor probability heatmap. The classification model considers patch-scale local features, and the segmentation model can take global information into account. We also propose a new optimization method that retains gradient information and trains the model partially for end-to-end learning with limited GPU memory capacity. We apply our method to the tumor/normal prediction on WSIs and the classification performance is improved compared with the conventional patch-based method.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge