Kengo Araki
Cluster Entropy: Active Domain Adaptation in Pathological Image Segmentation
Apr 26, 2023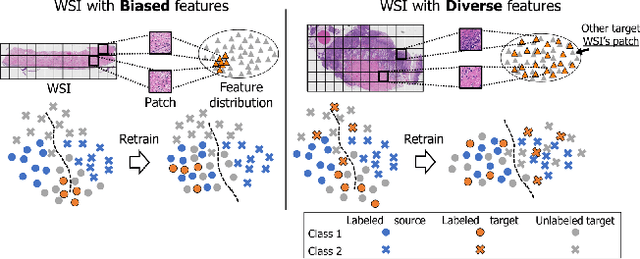
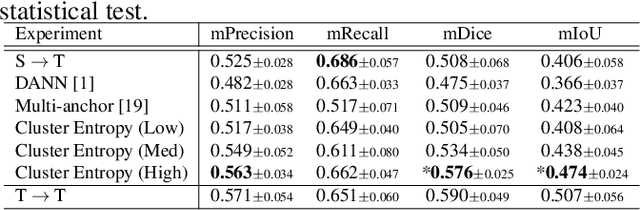

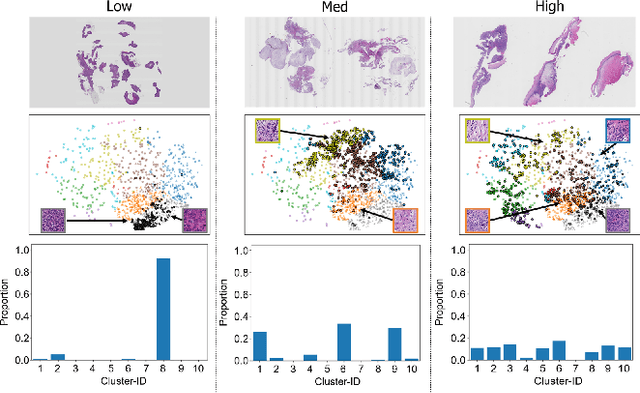
Abstract:The domain shift in pathological segmentation is an important problem, where a network trained by a source domain (collected at a specific hospital) does not work well in the target domain (from different hospitals) due to the different image features. Due to the problems of class imbalance and different class prior of pathology, typical unsupervised domain adaptation methods do not work well by aligning the distribution of source domain and target domain. In this paper, we propose a cluster entropy for selecting an effective whole slide image (WSI) that is used for semi-supervised domain adaptation. This approach can measure how the image features of the WSI cover the entire distribution of the target domain by calculating the entropy of each cluster and can significantly improve the performance of domain adaptation. Our approach achieved competitive results against the prior arts on datasets collected from two hospitals.
Cluster-Guided Semi-Supervised Domain Adaptation for Imbalanced Medical Image Classification
Mar 02, 2023Abstract:Semi-supervised domain adaptation is a technique to build a classifier for a target domain by modifying a classifier in another (source) domain using many unlabeled samples and a small number of labeled samples from the target domain. In this paper, we develop a semi-supervised domain adaptation method, which has robustness to class-imbalanced situations, which are common in medical image classification tasks. For robustness, we propose a weakly-supervised clustering pipeline to obtain high-purity clusters and utilize the clusters in representation learning for domain adaptation. The proposed method showed state-of-the-art performance in the experiment using severely class-imbalanced pathological image patches.
Patch-Based Cervical Cancer Segmentation using Distance from Boundary of Tissue
Aug 19, 2021

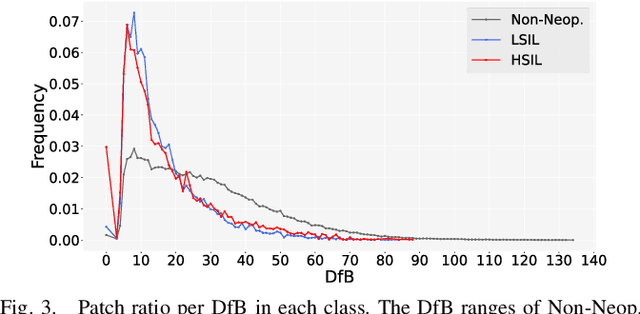

Abstract:Pathological diagnosis is used for examining cancer in detail, and its automation is in demand. To automatically segment each cancer area, a patch-based approach is usually used since a Whole Slide Image (WSI) is huge. However, this approach loses the global information needed to distinguish between classes. In this paper, we utilized the Distance from the Boundary of tissue (DfB), which is global information that can be extracted from the original image. We experimentally applied our method to the three-class classification of cervical cancer, and found that it improved the total performance compared with the conventional method.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge