Shota Harada
Domain Adaptation for Ulcerative Colitis Severity Estimation Using Patient-Level Diagnoses
Sep 18, 2025



Abstract:The development of methods to estimate the severity of Ulcerative Colitis (UC) is of significant importance. However, these methods often suffer from domain shifts caused by differences in imaging devices and clinical settings across hospitals. Although several domain adaptation methods have been proposed to address domain shift, they still struggle with the lack of supervision in the target domain or the high cost of annotation. To overcome these challenges, we propose a novel Weakly Supervised Domain Adaptation method that leverages patient-level diagnostic results, which are routinely recorded in UC diagnosis, as weak supervision in the target domain. The proposed method aligns class-wise distributions across domains using Shared Aggregation Tokens and a Max-Severity Triplet Loss, which leverages the characteristic that patient-level diagnoses are determined by the most severe region within each patient. Experimental results demonstrate that our method outperforms comparative DA approaches, improving UC severity estimation in a domain-shifted setting.
Domain Generalization of Pathological Image Segmentation by Patch-Level and WSI-Level Contrastive Learning
Aug 11, 2025


Abstract:In this paper, we address domain shifts in pathological images by focusing on shifts within whole slide images~(WSIs), such as patient characteristics and tissue thickness, rather than shifts between hospitals. Traditional approaches rely on multi-hospital data, but data collection challenges often make this impractical. Therefore, the proposed domain generalization method captures and leverages intra-hospital domain shifts by clustering WSI-level features from non-tumor regions and treating these clusters as domains. To mitigate domain shift, we apply contrastive learning to reduce feature gaps between WSI pairs from different clusters. The proposed method introduces a two-stage contrastive learning approach WSI-level and patch-level contrastive learning to minimize these gaps effectively.
Cluster Entropy: Active Domain Adaptation in Pathological Image Segmentation
Apr 26, 2023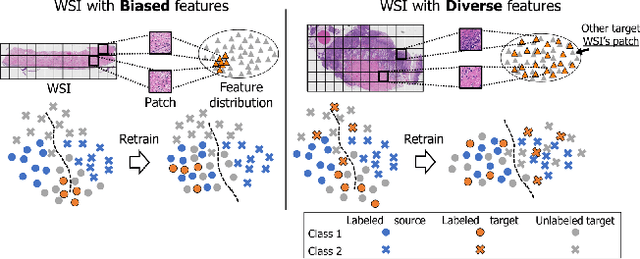
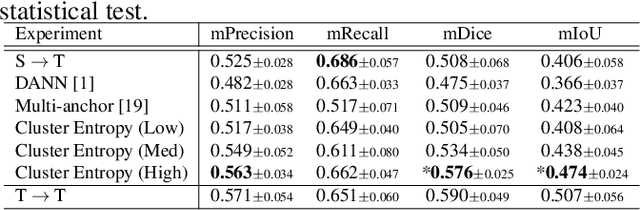

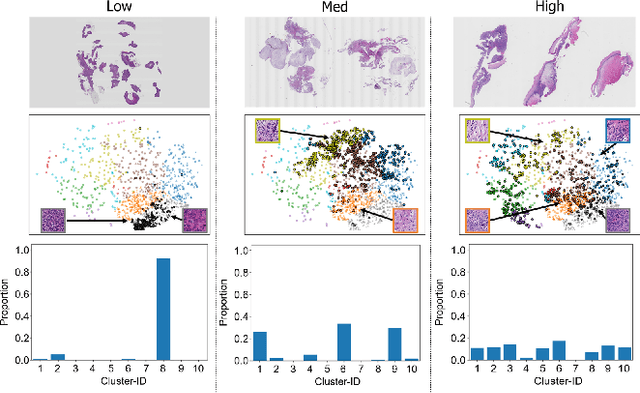
Abstract:The domain shift in pathological segmentation is an important problem, where a network trained by a source domain (collected at a specific hospital) does not work well in the target domain (from different hospitals) due to the different image features. Due to the problems of class imbalance and different class prior of pathology, typical unsupervised domain adaptation methods do not work well by aligning the distribution of source domain and target domain. In this paper, we propose a cluster entropy for selecting an effective whole slide image (WSI) that is used for semi-supervised domain adaptation. This approach can measure how the image features of the WSI cover the entire distribution of the target domain by calculating the entropy of each cluster and can significantly improve the performance of domain adaptation. Our approach achieved competitive results against the prior arts on datasets collected from two hospitals.
Cluster-Guided Semi-Supervised Domain Adaptation for Imbalanced Medical Image Classification
Mar 02, 2023Abstract:Semi-supervised domain adaptation is a technique to build a classifier for a target domain by modifying a classifier in another (source) domain using many unlabeled samples and a small number of labeled samples from the target domain. In this paper, we develop a semi-supervised domain adaptation method, which has robustness to class-imbalanced situations, which are common in medical image classification tasks. For robustness, we propose a weakly-supervised clustering pipeline to obtain high-purity clusters and utilize the clusters in representation learning for domain adaptation. The proposed method showed state-of-the-art performance in the experiment using severely class-imbalanced pathological image patches.
Order-Guided Disentangled Representation Learning for Ulcerative Colitis Classification with Limited Labels
Nov 06, 2021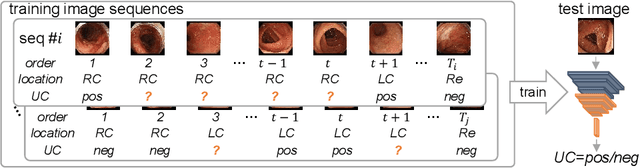
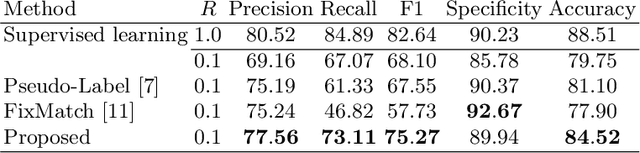

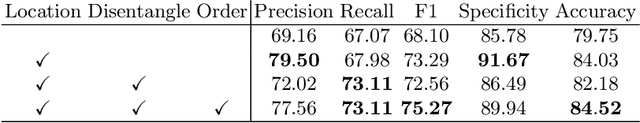
Abstract:Ulcerative colitis (UC) classification, which is an important task for endoscopic diagnosis, involves two main difficulties. First, endoscopic images with the annotation about UC (positive or negative) are usually limited. Second, they show a large variability in their appearance due to the location in the colon. Especially, the second difficulty prevents us from using existing semi-supervised learning techniques, which are the common remedy for the first difficulty. In this paper, we propose a practical semi-supervised learning method for UC classification by newly exploiting two additional features, the location in a colon (e.g., left colon) and image capturing order, both of which are often attached to individual images in endoscopic image sequences. The proposed method can extract the essential information of UC classification efficiently by a disentanglement process with those features. Experimental results demonstrate that the proposed method outperforms several existing semi-supervised learning methods in the classification task, even with a small number of annotated images.
Character-independent font identification
Jan 24, 2020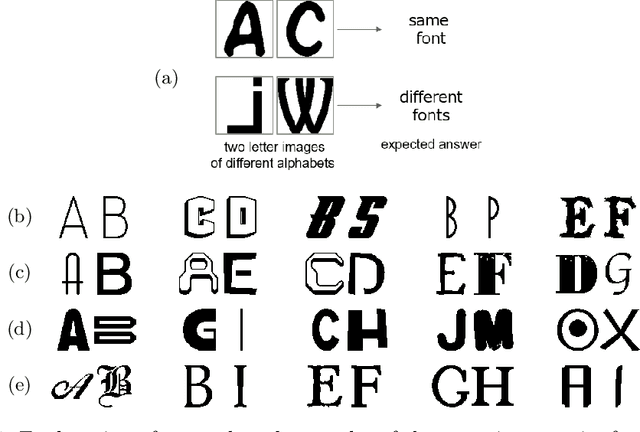
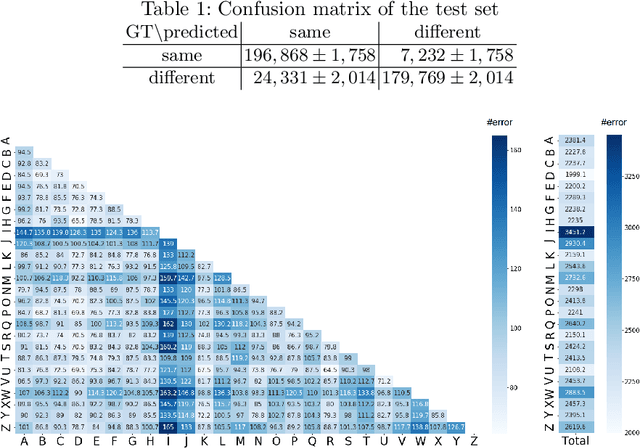
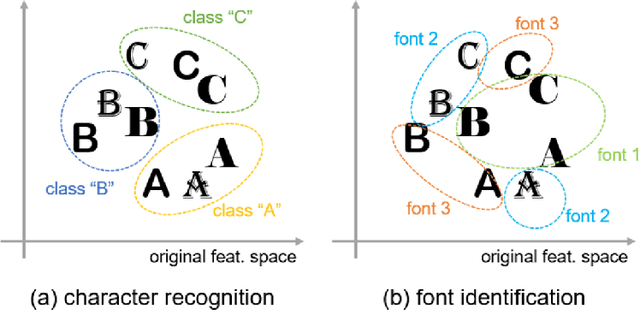
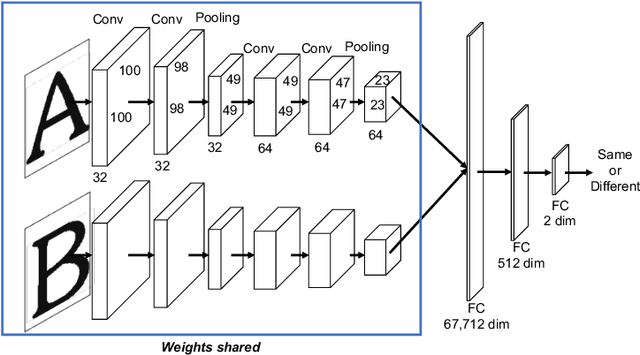
Abstract:There are a countless number of fonts with various shapes and styles. In addition, there are many fonts that only have subtle differences in features. Due to this, font identification is a difficult task. In this paper, we propose a method of determining if any two characters are from the same font or not. This is difficult due to the difference between fonts typically being smaller than the difference between alphabet classes. Additionally, the proposed method can be used with fonts regardless of whether they exist in the training or not. In order to accomplish this, we use a Convolutional Neural Network (CNN) trained with various font image pairs. In the experiment, the network is trained on image pairs of various fonts. We then evaluate the model on a different set of fonts that are unseen by the network. The evaluation is performed with an accuracy of 92.27%. Moreover, we analyzed the relationship between character classes and font identification accuracy.
Biosignal Generation and Latent Variable Analysis with Recurrent Generative Adversarial Networks
May 17, 2019



Abstract:The effectiveness of biosignal generation and data augmentation with biosignal generative models based on generative adversarial networks (GANs), which are a type of deep learning technique, was demonstrated in our previous paper. GAN-based generative models only learn the projection between a random distribution as input data and the distribution of training data.Therefore, the relationship between input and generated data is unclear, and the characteristics of the data generated from this model cannot be controlled. This study proposes a method for generating time-series data based on GANs and explores their ability to generate biosignals with certain classes and characteristics. Moreover, in the proposed method, latent variables are analyzed using canonical correlation analysis (CCA) to represent the relationship between input and generated data as canonical loadings. Using these loadings, we can control the characteristics of the data generated by the proposed method. The influence of class labels on generated data is analyzed by feeding the data interpolated between two class labels into the generator of the proposed GANs. The CCA of the latent variables is shown to be an effective method of controlling the generated data characteristics. We are able to model the distribution of the time-series data without requiring domain-dependent knowledge using the proposed method. Furthermore, it is possible to control the characteristics of these data by analyzing the model trained using the proposed method. To the best of our knowledge, this work is the first to generate biosignals using GANs while controlling the characteristics of the generated data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge