Masashi Fukayama
Domain Adaptive Multiple Instance Learning for Instance-level Prediction of Pathological Images
Apr 07, 2023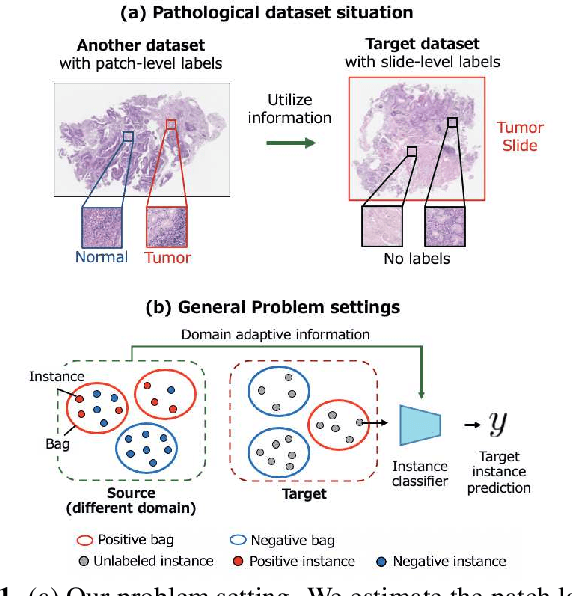
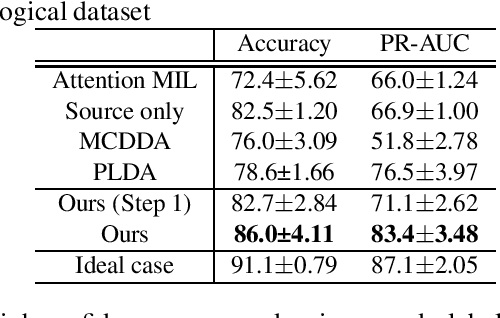
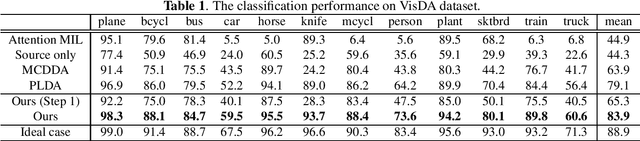
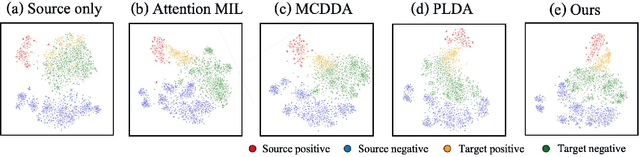
Abstract:Pathological image analysis is an important process for detecting abnormalities such as cancer from cell images. However, since the image size is generally very large, the cost of providing detailed annotations is high, which makes it difficult to apply machine learning techniques. One way to improve the performance of identifying abnormalities while keeping the annotation cost low is to use only labels for each slide, or to use information from another dataset that has already been labeled. However, such weak supervisory information often does not provide sufficient performance. In this paper, we proposed a new task setting to improve the classification performance of the target dataset without increasing annotation costs. And to solve this problem, we propose a pipeline that uses multiple instance learning (MIL) and domain adaptation (DA) methods. Furthermore, in order to combine the supervisory information of both methods effectively, we propose a method to create pseudo-labels with high confidence. We conducted experiments on the pathological image dataset we created for this study and showed that the proposed method significantly improves the classification performance compared to existing methods.
Multi-Stage Pathological Image Classification using Semantic Segmentation
Oct 10, 2019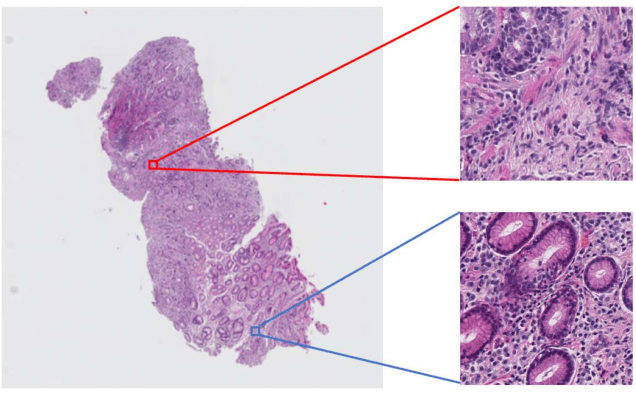
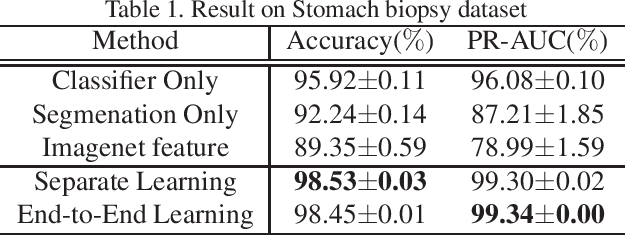
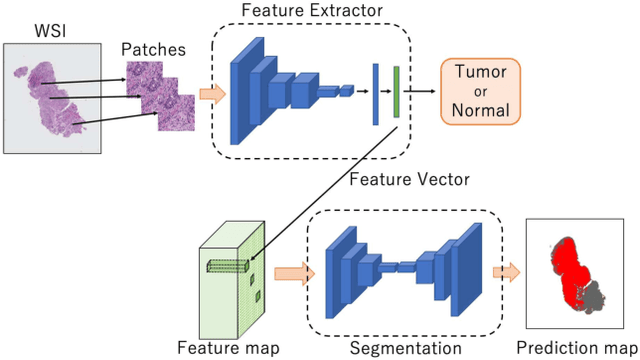
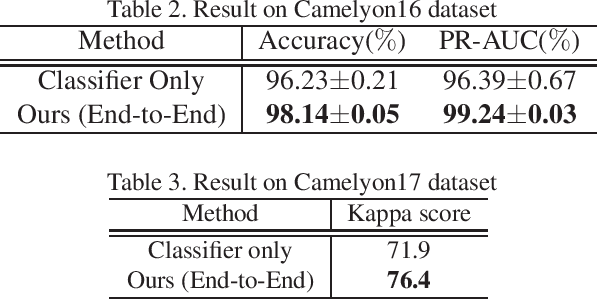
Abstract:Histopathological image analysis is an essential process for the discovery of diseases such as cancer. However, it is challenging to train CNN on whole slide images (WSIs) of gigapixel resolution considering the available memory capacity. Most of the previous works divide high resolution WSIs into small image patches and separately input them into the model to classify it as a tumor or a normal tissue. However, patch-based classification uses only patch-scale local information but ignores the relationship between neighboring patches. If we consider the relationship of neighboring patches and global features, we can improve the classification performance. In this paper, we propose a new model structure combining the patch-based classification model and whole slide-scale segmentation model in order to improve the prediction performance of automatic pathological diagnosis. We extract patch features from the classification model and input them into the segmentation model to obtain a whole slide tumor probability heatmap. The classification model considers patch-scale local features, and the segmentation model can take global information into account. We also propose a new optimization method that retains gradient information and trains the model partially for end-to-end learning with limited GPU memory capacity. We apply our method to the tumor/normal prediction on WSIs and the classification performance is improved compared with the conventional patch-based method.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge