Kazuya Nishimura
Towards Spatial Transcriptomics-guided Pathological Image Recognition with Batch-Agnostic Encoder
Mar 10, 2025



Abstract:Spatial transcriptomics (ST) is a novel technique that simultaneously captures pathological images and gene expression profiling with spatial coordinates. Since ST is closely related to pathological features such as disease subtypes, it may be valuable to augment image representation with pathological information. However, there are no attempts to leverage ST for image recognition ({\it i.e,} patch-level classification of subtypes of pathological image.). One of the big challenges is significant batch effects in spatial transcriptomics that make it difficult to extract pathological features of images from ST. In this paper, we propose a batch-agnostic contrastive learning framework that can extract consistent signals from gene expression of ST in multiple patients. To extract consistent signals from ST, we utilize the batch-agnostic gene encoder that is trained in a variational inference manner. Experiments demonstrated the effectiveness of our framework on a publicly available dataset. Code is publicly available at https://github.com/naivete5656/TPIRBAE
Ordinal Multiple-instance Learning for Ulcerative Colitis Severity Estimation with Selective Aggregated Transformer
Nov 22, 2024



Abstract:Patient-level diagnosis of severity in ulcerative colitis (UC) is common in real clinical settings, where the most severe score in a patient is recorded. However, previous UC classification methods (i.e., image-level estimation) mainly assumed the input was a single image. Thus, these methods can not utilize severity labels recorded in real clinical settings. In this paper, we propose a patient-level severity estimation method by a transformer with selective aggregator tokens, where a severity label is estimated from multiple images taken from a patient, similar to a clinical setting. Our method can effectively aggregate features of severe parts from a set of images captured in each patient, and it facilitates improving the discriminative ability between adjacent severity classes. Experiments demonstrate the effectiveness of the proposed method on two datasets compared with the state-of-the-art MIL methods. Moreover, we evaluated our method in real clinical settings and confirmed that our method outperformed the previous image-level methods. The code is publicly available at https://github.com/Shiku-Kaito/Ordinal-Multiple-instance-Learning-for-Ulcerative-Colitis-Severity-Estimation.
Proportion Estimation by Masked Learning from Label Proportion
May 08, 2024Abstract:The PD-L1 rate, the number of PD-L1 positive tumor cells over the total number of all tumor cells, is an important metric for immunotherapy. This metric is recorded as diagnostic information with pathological images. In this paper, we propose a proportion estimation method with a small amount of cell-level annotation and proportion annotation, which can be easily collected. Since the PD-L1 rate is calculated from only `tumor cells' and not using `non-tumor cells', we first detect tumor cells with a detection model. Then, we estimate the PD-L1 proportion by introducing a masking technique to `learning from label proportion.' In addition, we propose a weighted focal proportion loss to address data imbalance problems. Experiments using clinical data demonstrate the effectiveness of our method. Our method achieved the best performance in comparisons.
Mitosis Detection from Partial Annotation by Dataset Generation via Frame-Order Flipping
Jul 09, 2023



Abstract:Detection of mitosis events plays an important role in biomedical research. Deep-learning-based mitosis detection methods have achieved outstanding performance with a certain amount of labeled data. However, these methods require annotations for each imaging condition. Collecting labeled data involves time-consuming human labor. In this paper, we propose a mitosis detection method that can be trained with partially annotated sequences. The base idea is to generate a fully labeled dataset from the partial labels and train a mitosis detection model with the generated dataset. First, we generate an image pair not containing mitosis events by frame-order flipping. Then, we paste mitosis events to the image pair by alpha-blending pasting and generate a fully labeled dataset. We demonstrate the performance of our method on four datasets, and we confirm that our method outperforms other comparisons which use partially labeled sequences.
Effective Pseudo-Labeling based on Heatmap for Unsupervised Domain Adaptation in Cell Detection
Mar 09, 2023



Abstract:Cell detection is an important task in biomedical research. Recently, deep learning methods have made it possible to improve the performance of cell detection. However, a detection network trained with training data under a specific condition (source domain) may not work well on data under other conditions (target domains), which is called the domain shift problem. In particular, cells are cultured under different conditions depending on the purpose of the research. Characteristics, e.g., the shapes and density of the cells, change depending on the conditions, and such changes may cause domain shift problems. Here, we propose an unsupervised domain adaptation method for cell detection using a pseudo-cell-position heatmap, where the cell centroid is at the peak of a Gaussian distribution in the map and selective pseudo-labeling. In the prediction result for the target domain, even if the peak location is correct, the signal distribution around the peak often has a non-Gaussian shape. The pseudo-cell-position heatmap is thus re-generated using the peak positions in the predicted heatmap to have a clear Gaussian shape. Our method selects confident pseudo-cell-position heatmaps based on uncertainty and curriculum learning. We conducted numerous experiments showing that, compared with the existing methods, our method improved detection performance under different conditions.
* 16 pages, 18 figures, Accepted in Medical Image Analysis 2022
Cell Detection from Imperfect Annotation by Pseudo Label Selection Using P-classification
Jul 21, 2021
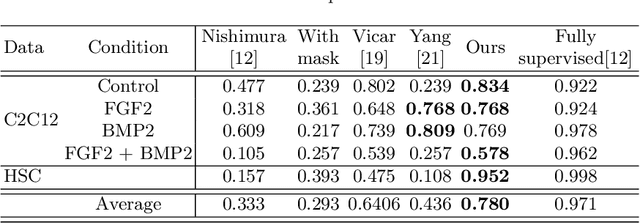
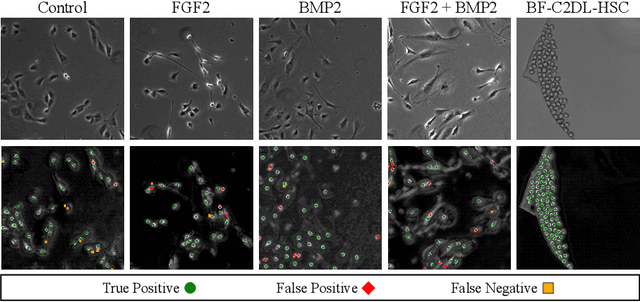
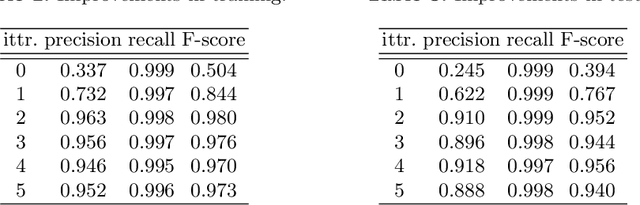
Abstract:Cell detection is an essential task in cell image analysis. Recent deep learning-based detection methods have achieved very promising results. In general, these methods require exhaustively annotating the cells in an entire image. If some of the cells are not annotated (imperfect annotation), the detection performance significantly degrades due to noisy labels. This often occurs in real collaborations with biologists and even in public data-sets. Our proposed method takes a pseudo labeling approach for cell detection from imperfect annotated data. A detection convolutional neural network (CNN) trained using such missing labeled data often produces over-detection. We treat partially labeled cells as positive samples and the detected positions except for the labeled cell as unlabeled samples. Then we select reliable pseudo labels from unlabeled data using recent machine learning techniques; positive-and-unlabeled (PU) learning and P-classification. Experiments using microscopy images for five different conditions demonstrate the effectiveness of the proposed method.
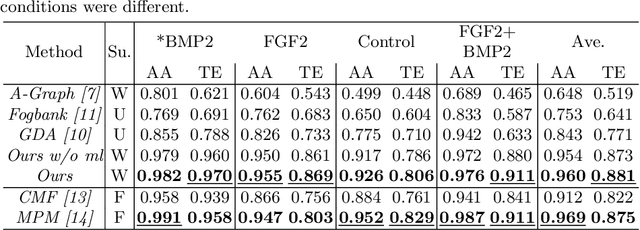
Cell Detection in Domain Shift Problem Using Pseudo-Cell-Position Heatmap
Jul 19, 2021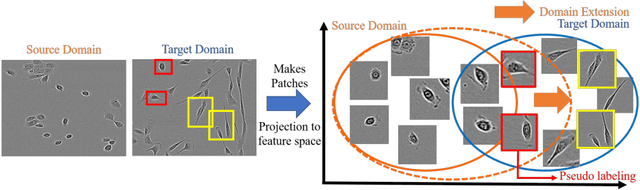

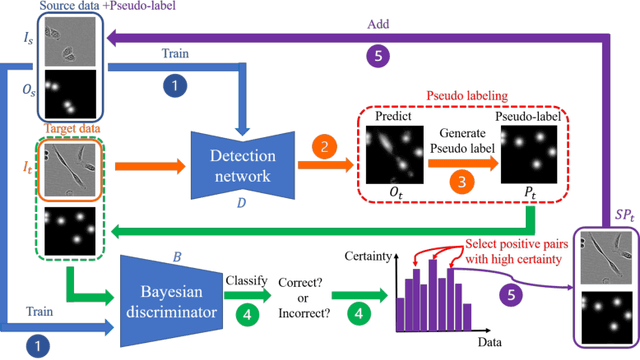
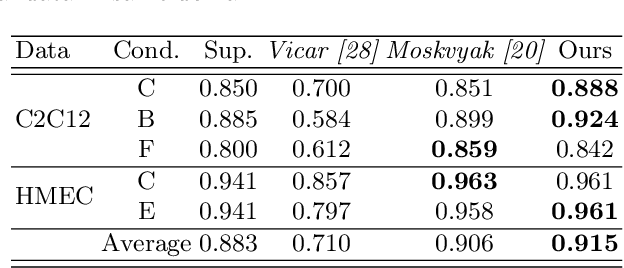
Abstract:The domain shift problem is an important issue in automatic cell detection. A detection network trained with training data under a specific condition (source domain) may not work well in data under other conditions (target domain). We propose an unsupervised domain adaptation method for cell detection using the pseudo-cell-position heatmap, where a cell centroid becomes a peak with a Gaussian distribution in the map. In the prediction result for the target domain, even if a peak location is correct, the signal distribution around the peak often has anon-Gaussian shape. The pseudo-cell-position heatmap is re-generated using the peak positions in the predicted heatmap to have a clear Gaussian shape. Our method selects confident pseudo-cell-position heatmaps using a Bayesian network and adds them to the training data in the next iteration. The method can incrementally extend the domain from the source domain to the target domain in a semi-supervised manner. In the experiments using 8 combinations of domains, the proposed method outperformed the existing domain adaptation methods.
Semi-supervised Cell Detection in Time-lapse Images Using Temporal Consistency
Jul 19, 2021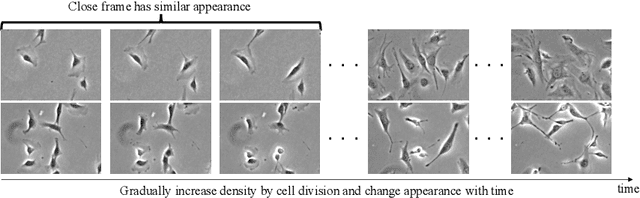
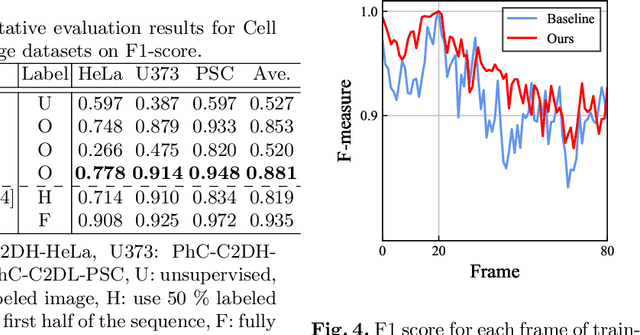

Abstract:Cell detection is the task of detecting the approximate positions of cell centroids from microscopy images. Recently, convolutional neural network-based approaches have achieved promising performance. However, these methods require a certain amount of annotation for each imaging condition. This annotation is a time-consuming and labor-intensive task. To overcome this problem, we propose a semi-supervised cell-detection method that effectively uses a time-lapse sequence with one labeled image and the other images unlabeled. First, we train a cell-detection network with a one-labeled image and estimate the unlabeled images with the trained network. We then select high-confidence positions from the estimations by tracking the detected cells from the labeled frame to those far from it. Next, we generate pseudo-labels from the tracking results and train the network by using pseudo-labels. We evaluated our method for seven conditions of public datasets, and we achieved the best results relative to other semi-supervised methods. Our code is available at https://github.com/naivete5656/SCDTC
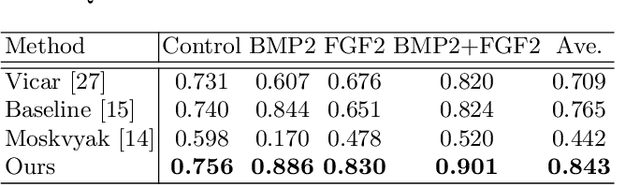
Weakly-Supervised Cell Tracking via Backward-and-Forward Propagation
Jul 30, 2020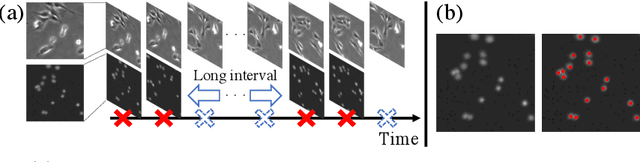
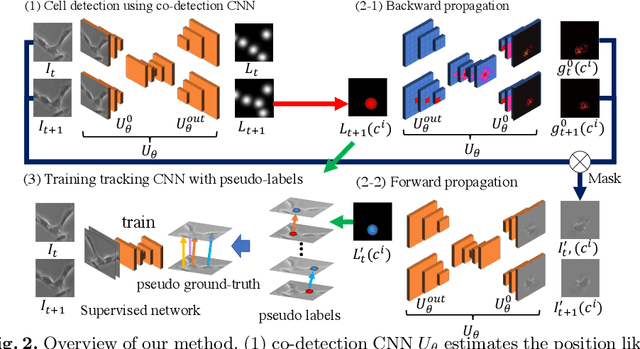
Abstract:We propose a weakly-supervised cell tracking method that can train a convolutional neural network (CNN) by using only the annotation of "cell detection" (i.e., the coordinates of cell positions) without association information, in which cell positions can be easily obtained by nuclear staining. First, we train co-detection CNN that detects cells in successive frames by using weak-labels. Our key assumption is that co-detection CNN implicitly learns association in addition to detection. To obtain the association, we propose a backward-and-forward propagation method that analyzes the correspondence of cell positions in the outputs of co-detection CNN. Experiments demonstrated that the proposed method can associate cells by analyzing co-detection CNN. Even though the method uses only weak supervision, the performance of our method was almost the same as the state-of-the-art supervised method. Code is publicly available in https://github.com/naivete5656/WSCTBFP
Spatial-Temporal Mitosis Detection in Phase-Contrast Microscopy via Likelihood Map Estimation by 3DCNN
Jun 01, 2020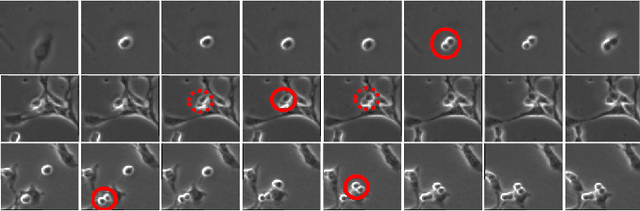
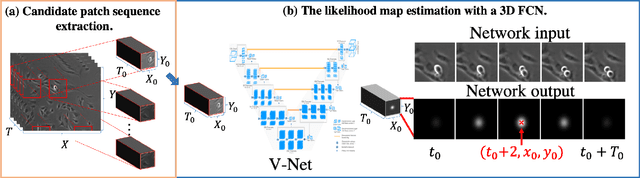
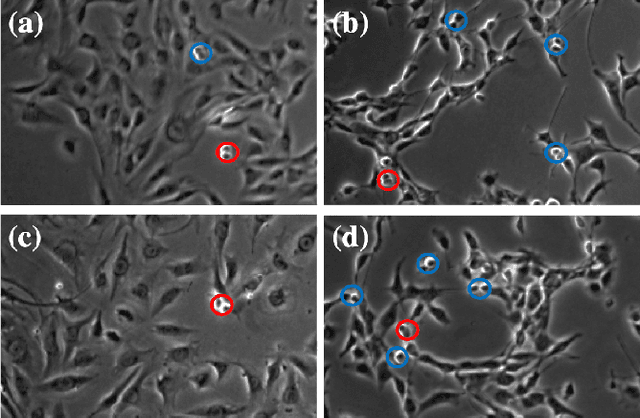

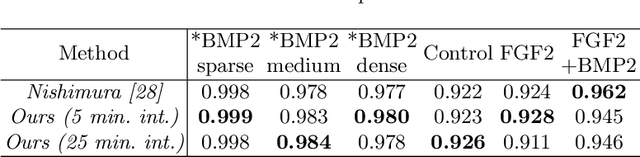
Abstract:Automated mitotic detection in time-lapse phasecontrast microscopy provides us much information for cell behavior analysis, and thus several mitosis detection methods have been proposed. However, these methods still have two problems; 1) they cannot detect multiple mitosis events when there are closely placed. 2) they do not consider the annotation gaps, which may occur since the appearances of mitosis cells are very similar before and after the annotated frame. In this paper, we propose a novel mitosis detection method that can detect multiple mitosis events in a candidate sequence and mitigate the human annotation gap via estimating a spatiotemporal likelihood map by 3DCNN. In this training, the loss gradually decreases with the gap size between ground truth and estimation. This mitigates the annotation gaps. Our method outperformed the compared methods in terms of F1- score using a challenging dataset that contains the data under four different conditions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge