Junya Hayashida
Weakly-Supervised Cell Tracking via Backward-and-Forward Propagation
Jul 30, 2020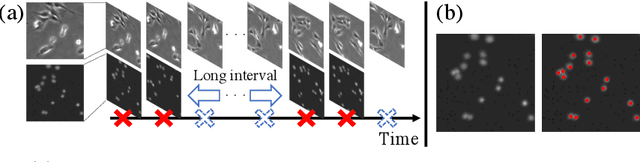
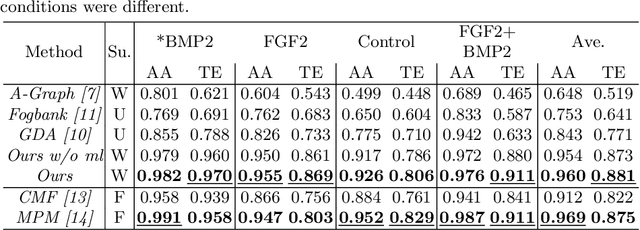
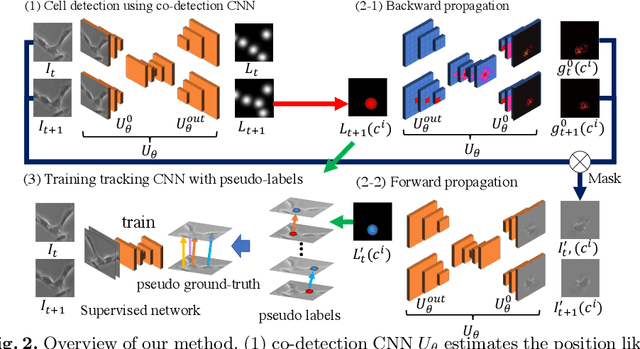
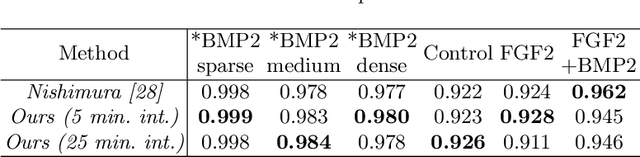
Abstract:We propose a weakly-supervised cell tracking method that can train a convolutional neural network (CNN) by using only the annotation of "cell detection" (i.e., the coordinates of cell positions) without association information, in which cell positions can be easily obtained by nuclear staining. First, we train co-detection CNN that detects cells in successive frames by using weak-labels. Our key assumption is that co-detection CNN implicitly learns association in addition to detection. To obtain the association, we propose a backward-and-forward propagation method that analyzes the correspondence of cell positions in the outputs of co-detection CNN. Experiments demonstrated that the proposed method can associate cells by analyzing co-detection CNN. Even though the method uses only weak supervision, the performance of our method was almost the same as the state-of-the-art supervised method. Code is publicly available in https://github.com/naivete5656/WSCTBFP
MPM: Joint Representation of Motion and Position Map for Cell Tracking
Feb 26, 2020
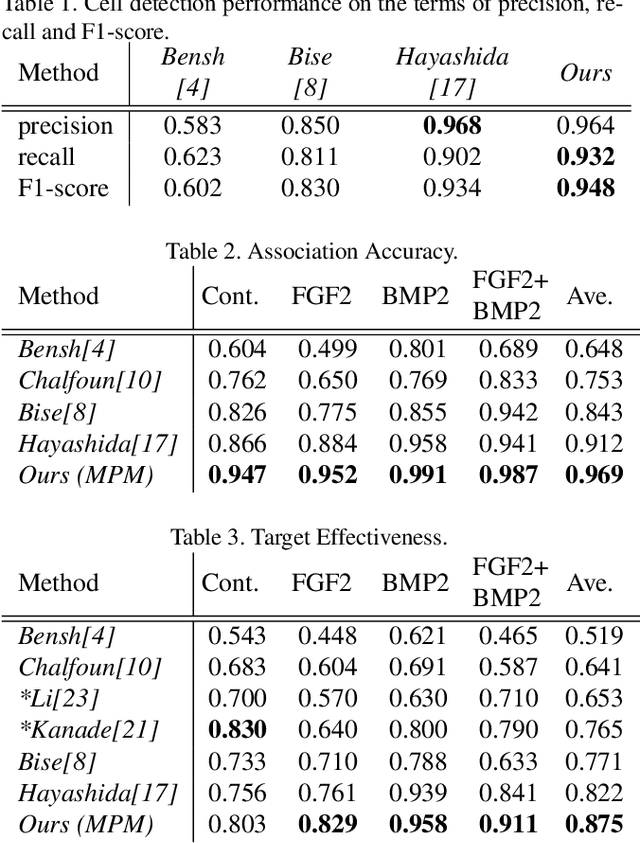
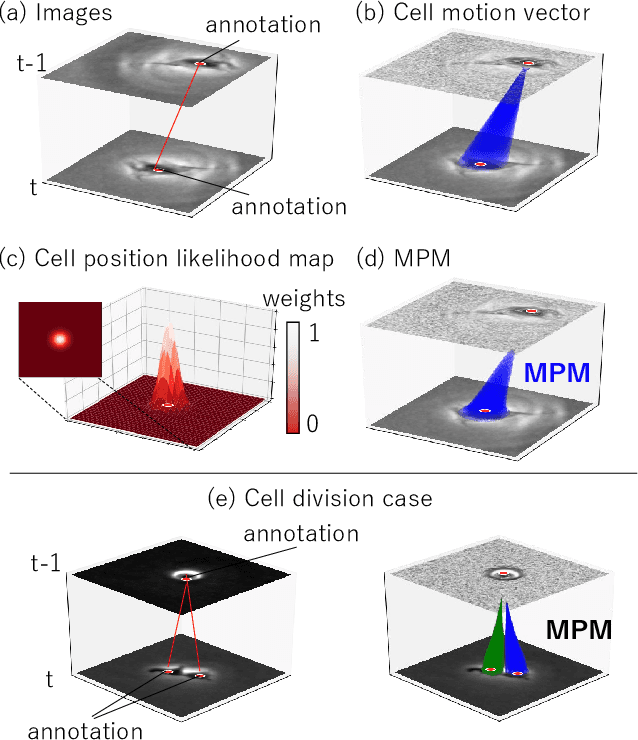

Abstract:Conventional cell tracking methods detect multiple cells in each frame (detection) and then associate the detection results in successive time-frames (association). Most cell tracking methods perform the association task independently from the detection task. However, there is no guarantee of preserving coherence between these tasks, and lack of coherence may adversely affect tracking performance. In this paper, we propose the Motion and Position Map (MPM) that jointly represents both detection and association for not only migration but also cell division. It guarantees coherence such that if a cell is detected, the corresponding motion flow can always be obtained. It is a simple but powerful method for multi-object tracking in dense environments. We compared the proposed method with current tracking methods under various conditions in real biological images and found that it outperformed the state-of-the-art (+5.2\% improvement compared to the second-best).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge