Zheming Li
Kimi K2.5: Visual Agentic Intelligence
Feb 02, 2026Abstract:We introduce Kimi K2.5, an open-source multimodal agentic model designed to advance general agentic intelligence. K2.5 emphasizes the joint optimization of text and vision so that two modalities enhance each other. This includes a series of techniques such as joint text-vision pre-training, zero-vision SFT, and joint text-vision reinforcement learning. Building on this multimodal foundation, K2.5 introduces Agent Swarm, a self-directed parallel agent orchestration framework that dynamically decomposes complex tasks into heterogeneous sub-problems and executes them concurrently. Extensive evaluations show that Kimi K2.5 achieves state-of-the-art results across various domains including coding, vision, reasoning, and agentic tasks. Agent Swarm also reduces latency by up to $4.5\times$ over single-agent baselines. We release the post-trained Kimi K2.5 model checkpoint to facilitate future research and real-world applications of agentic intelligence.
Kimi K2: Open Agentic Intelligence
Jul 28, 2025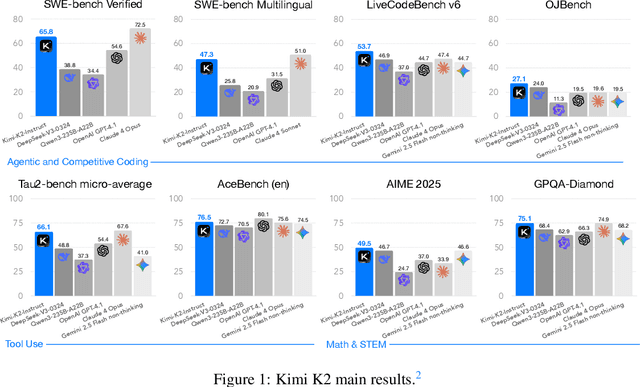

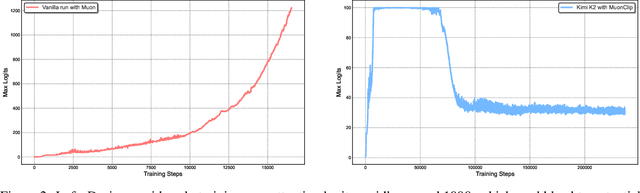
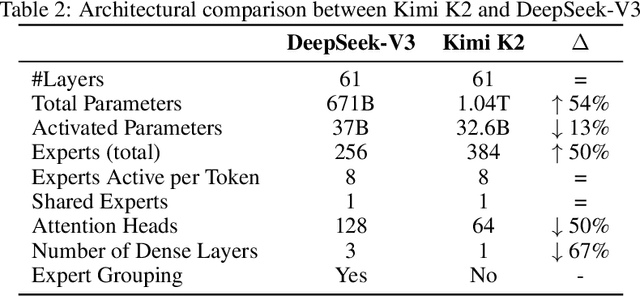
Abstract:We introduce Kimi K2, a Mixture-of-Experts (MoE) large language model with 32 billion activated parameters and 1 trillion total parameters. We propose the MuonClip optimizer, which improves upon Muon with a novel QK-clip technique to address training instability while enjoying the advanced token efficiency of Muon. Based on MuonClip, K2 was pre-trained on 15.5 trillion tokens with zero loss spike. During post-training, K2 undergoes a multi-stage post-training process, highlighted by a large-scale agentic data synthesis pipeline and a joint reinforcement learning (RL) stage, where the model improves its capabilities through interactions with real and synthetic environments. Kimi K2 achieves state-of-the-art performance among open-source non-thinking models, with strengths in agentic capabilities. Notably, K2 obtains 66.1 on Tau2-Bench, 76.5 on ACEBench (En), 65.8 on SWE-Bench Verified, and 47.3 on SWE-Bench Multilingual -- surpassing most open and closed-sourced baselines in non-thinking settings. It also exhibits strong capabilities in coding, mathematics, and reasoning tasks, with a score of 53.7 on LiveCodeBench v6, 49.5 on AIME 2025, 75.1 on GPQA-Diamond, and 27.1 on OJBench, all without extended thinking. These results position Kimi K2 as one of the most capable open-source large language models to date, particularly in software engineering and agentic tasks. We release our base and post-trained model checkpoints to facilitate future research and applications of agentic intelligence.
Mooncake: A KVCache-centric Disaggregated Architecture for LLM Serving
Jul 02, 2024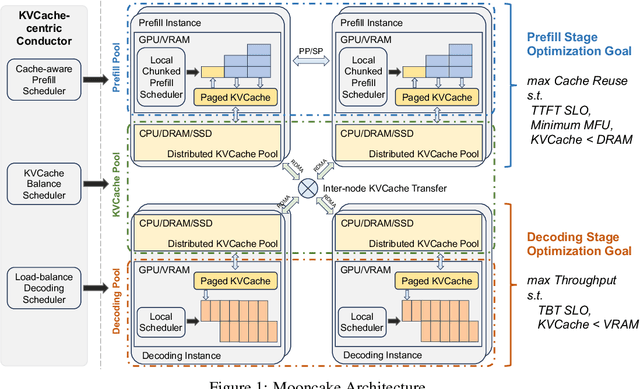
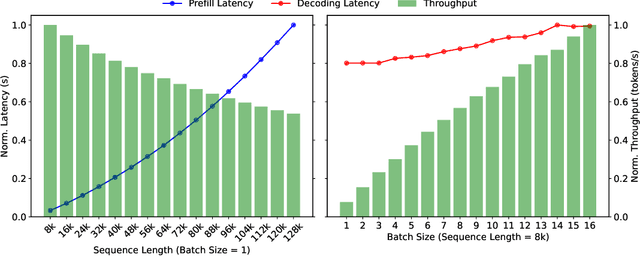

Abstract:Mooncake is the serving platform for Kimi, a leading LLM service provided by Moonshot AI. It features a KVCache-centric disaggregated architecture that separates the prefill and decoding clusters. It also leverages the underutilized CPU, DRAM, and SSD resources of the GPU cluster to implement a disaggregated cache of KVCache. The core of Mooncake is its KVCache-centric scheduler, which balances maximizing overall effective throughput while meeting latency-related Service Level Objectives (SLOs). Unlike traditional studies that assume all requests will be processed, Mooncake faces challenges due to highly overloaded scenarios. To mitigate these, we developed a prediction-based early rejection policy. Experiments show that Mooncake excels in long-context scenarios. Compared to the baseline method, Mooncake can achieve up to a 525% increase in throughput in certain simulated scenarios while adhering to SLOs. Under real workloads, Mooncake's innovative architecture enables Kimi to handle 75% more requests.
Identification of Pediatric Respiratory Diseases Using Fine-grained Diagnosis System
Aug 24, 2021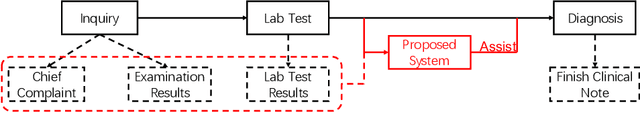
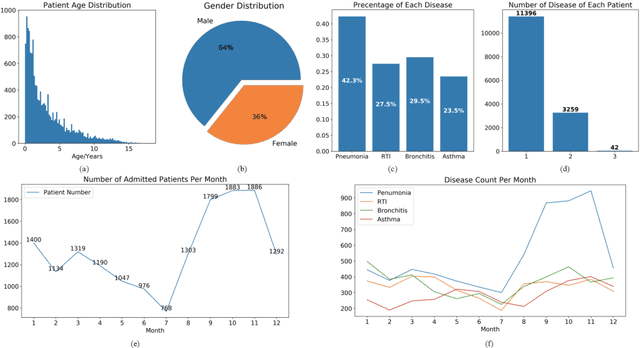
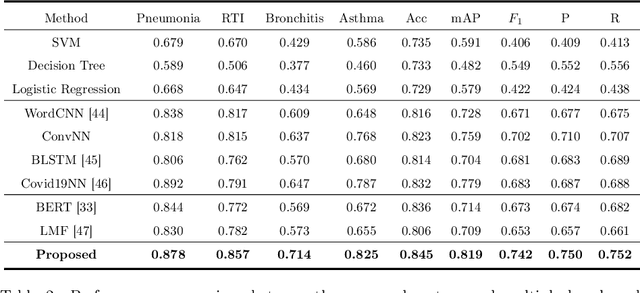
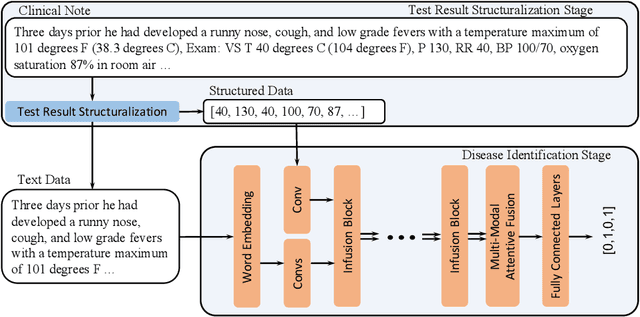
Abstract:Respiratory diseases, including asthma, bronchitis, pneumonia, and upper respiratory tract infection (RTI), are among the most common diseases in clinics. The similarities among the symptoms of these diseases precludes prompt diagnosis upon the patients' arrival. In pediatrics, the patients' limited ability in expressing their situation makes precise diagnosis even harder. This becomes worse in primary hospitals, where the lack of medical imaging devices and the doctors' limited experience further increase the difficulty of distinguishing among similar diseases. In this paper, a pediatric fine-grained diagnosis-assistant system is proposed to provide prompt and precise diagnosis using solely clinical notes upon admission, which would assist clinicians without changing the diagnostic process. The proposed system consists of two stages: a test result structuralization stage and a disease identification stage. The first stage structuralizes test results by extracting relevant numerical values from clinical notes, and the disease identification stage provides a diagnosis based on text-form clinical notes and the structured data obtained from the first stage. A novel deep learning algorithm was developed for the disease identification stage, where techniques including adaptive feature infusion and multi-modal attentive fusion were introduced to fuse structured and text data together. Clinical notes from over 12000 patients with respiratory diseases were used to train a deep learning model, and clinical notes from a non-overlapping set of about 1800 patients were used to evaluate the performance of the trained model. The average precisions (AP) for pneumonia, RTI, bronchitis and asthma are 0.878, 0.857, 0.714, and 0.825, respectively, achieving a mean AP (mAP) of 0.819.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge