Qiang Shu
TDFANet: Encoding Sequential 4D Radar Point Clouds Using Trajectory-Guided Deformable Feature Aggregation for Place Recognition
Apr 07, 2025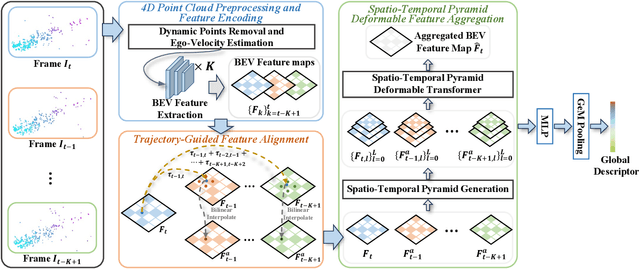
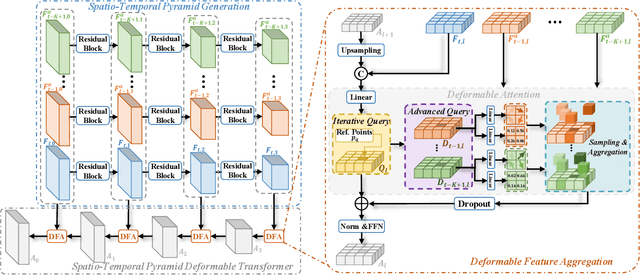
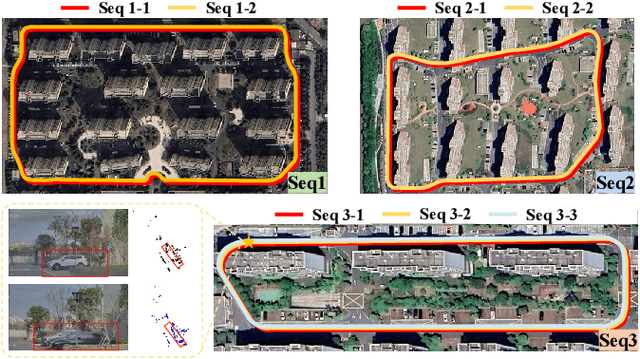
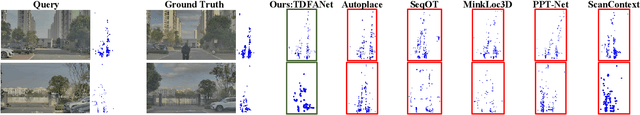
Abstract:Place recognition is essential for achieving closed-loop or global positioning in autonomous vehicles and mobile robots. Despite recent advancements in place recognition using 2D cameras or 3D LiDAR, it remains to be seen how to use 4D radar for place recognition - an increasingly popular sensor for its robustness against adverse weather and lighting conditions. Compared to LiDAR point clouds, radar data are drastically sparser, noisier and in much lower resolution, which hampers their ability to effectively represent scenes, posing significant challenges for 4D radar-based place recognition. This work addresses these challenges by leveraging multi-modal information from sequential 4D radar scans and effectively extracting and aggregating spatio-temporal features.Our approach follows a principled pipeline that comprises (1) dynamic points removal and ego-velocity estimation from velocity property, (2) bird's eye view (BEV) feature encoding on the refined point cloud, (3) feature alignment using BEV feature map motion trajectory calculated by ego-velocity, (4) multi-scale spatio-temporal features of the aligned BEV feature maps are extracted and aggregated.Real-world experimental results validate the feasibility of the proposed method and demonstrate its robustness in handling dynamic environments. Source codes are available.
Identification of Pediatric Respiratory Diseases Using Fine-grained Diagnosis System
Aug 24, 2021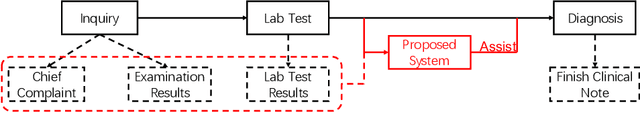
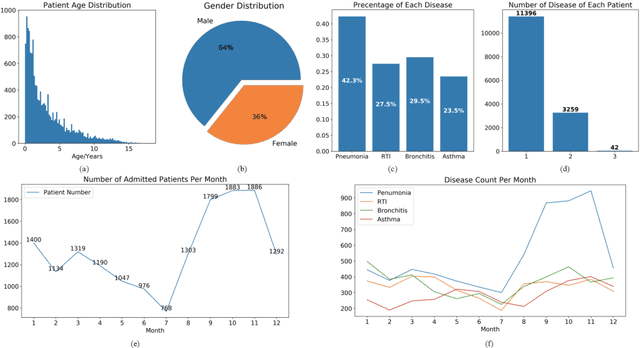
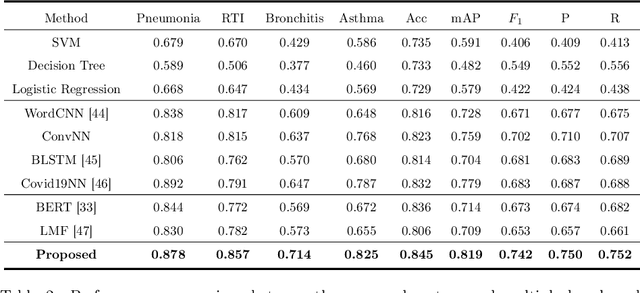
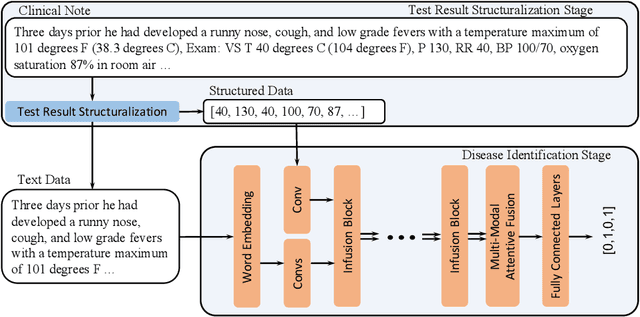
Abstract:Respiratory diseases, including asthma, bronchitis, pneumonia, and upper respiratory tract infection (RTI), are among the most common diseases in clinics. The similarities among the symptoms of these diseases precludes prompt diagnosis upon the patients' arrival. In pediatrics, the patients' limited ability in expressing their situation makes precise diagnosis even harder. This becomes worse in primary hospitals, where the lack of medical imaging devices and the doctors' limited experience further increase the difficulty of distinguishing among similar diseases. In this paper, a pediatric fine-grained diagnosis-assistant system is proposed to provide prompt and precise diagnosis using solely clinical notes upon admission, which would assist clinicians without changing the diagnostic process. The proposed system consists of two stages: a test result structuralization stage and a disease identification stage. The first stage structuralizes test results by extracting relevant numerical values from clinical notes, and the disease identification stage provides a diagnosis based on text-form clinical notes and the structured data obtained from the first stage. A novel deep learning algorithm was developed for the disease identification stage, where techniques including adaptive feature infusion and multi-modal attentive fusion were introduced to fuse structured and text data together. Clinical notes from over 12000 patients with respiratory diseases were used to train a deep learning model, and clinical notes from a non-overlapping set of about 1800 patients were used to evaluate the performance of the trained model. The average precisions (AP) for pneumonia, RTI, bronchitis and asthma are 0.878, 0.857, 0.714, and 0.825, respectively, achieving a mean AP (mAP) of 0.819.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge