Heshui Shi
GASNet: Weakly-supervised Framework for COVID-19 Lesion Segmentation
Oct 19, 2020



Abstract:Segmentation of infected areas in chest CT volumes is of great significance for further diagnosis and treatment of COVID-19 patients. Due to the complex shapes and varied appearances of lesions, a large number of voxel-level labeled samples are generally required to train a lesion segmentation network, which is a main bottleneck for developing deep learning based medical image segmentation algorithms. In this paper, we propose a weakly-supervised lesion segmentation framework by embedding the Generative Adversarial training process into the Segmentation Network, which is called GASNet. GASNet is optimized to segment the lesion areas of a COVID-19 CT by the segmenter, and to replace the abnormal appearance with a generated normal appearance by the generator, so that the restored CT volumes are indistinguishable from healthy CT volumes by the discriminator. GASNet is supervised by chest CT volumes of many healthy and COVID-19 subjects without voxel-level annotations. Experiments on three public databases show that when using as few as one voxel-level labeled sample, the performance of GASNet is comparable to fully-supervised segmentation algorithms trained on dozens of voxel-level labeled samples.
Learning Directional Feature Maps for Cardiac MRI Segmentation
Jul 22, 2020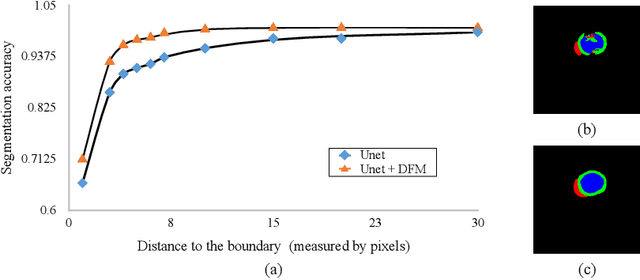
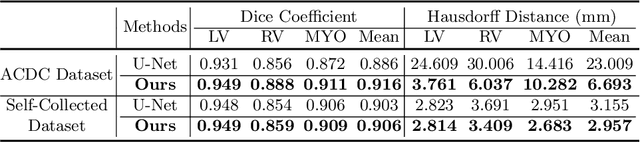
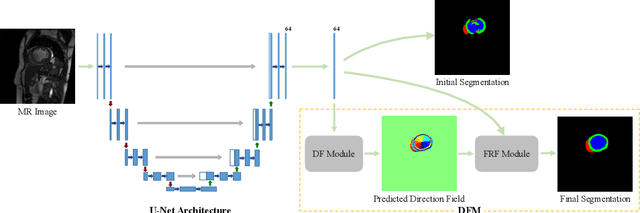
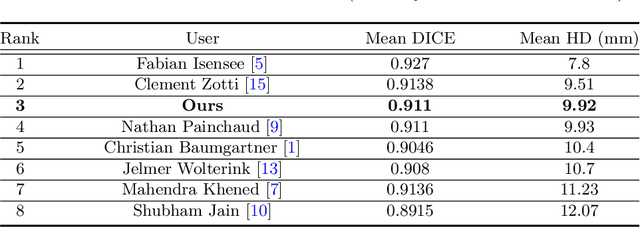
Abstract:Cardiac MRI segmentation plays a crucial role in clinical diagnosis for evaluating personalized cardiac performance parameters. Due to the indistinct boundaries and heterogeneous intensity distributions in the cardiac MRI, most existing methods still suffer from two aspects of challenges: inter-class indistinction and intra-class inconsistency. To tackle these two problems, we propose a novel method to exploit the directional feature maps, which can simultaneously strengthen the differences between classes and the similarities within classes. Specifically, we perform cardiac segmentation and learn a direction field pointing away from the nearest cardiac tissue boundary to each pixel via a direction field (DF) module. Based on the learned direction field, we then propose a feature rectification and fusion (FRF) module to improve the original segmentation features, and obtain the final segmentation. The proposed modules are simple yet effective and can be flexibly added to any existing segmentation network without excessively increasing time and space complexity. We evaluate the proposed method on the 2017 MICCAI Automated Cardiac Diagnosis Challenge (ACDC) dataset and a large-scale self-collected dataset, showing good segmentation performance and robust generalization ability of the proposed method.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge