Yongchao Xu
LIGM
IVC-Prune: Revealing the Implicit Visual Coordinates in LVLMs for Vision Token Pruning
Feb 03, 2026Abstract:Large Vision-Language Models (LVLMs) achieve impressive performance across multiple tasks. A significant challenge, however, is their prohibitive inference cost when processing high-resolution visual inputs. While visual token pruning has emerged as a promising solution, existing methods that primarily focus on semantic relevance often discard tokens that are crucial for spatial reasoning. We address this gap through a novel insight into \emph{how LVLMs process spatial reasoning}. Specifically, we reveal that LVLMs implicitly establish visual coordinate systems through Rotary Position Embeddings (RoPE), where specific token positions serve as \textbf{implicit visual coordinates} (IVC tokens) that are essential for spatial reasoning. Based on this insight, we propose \textbf{IVC-Prune}, a training-free, prompt-aware pruning strategy that retains both IVC tokens and semantically relevant foreground tokens. IVC tokens are identified by theoretically analyzing the mathematical properties of RoPE, targeting positions at which its rotation matrices approximate identity matrix or the $90^\circ$ rotation matrix. Foreground tokens are identified through a robust two-stage process: semantic seed discovery followed by contextual refinement via value-vector similarity. Extensive evaluations across four representative LVLMs and twenty diverse benchmarks show that IVC-Prune reduces visual tokens by approximately 50\% while maintaining $\geq$ 99\% of the original performance and even achieving improvements on several benchmarks. Source codes are available at https://github.com/FireRedTeam/IVC-Prune.
A DeepSeek-Powered AI System for Automated Chest Radiograph Interpretation in Clinical Practice
Dec 23, 2025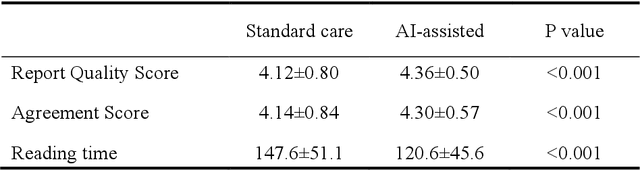
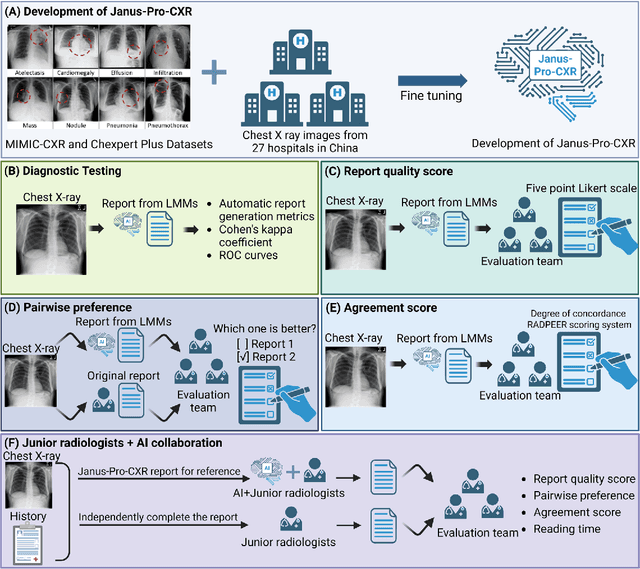
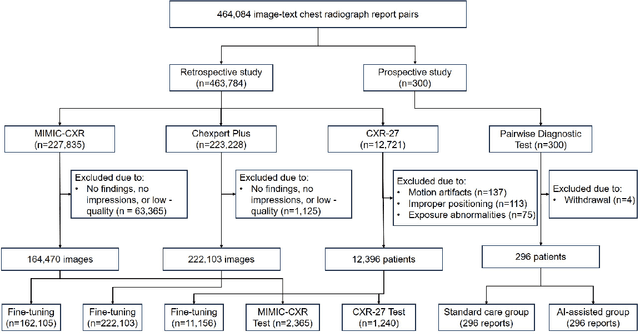
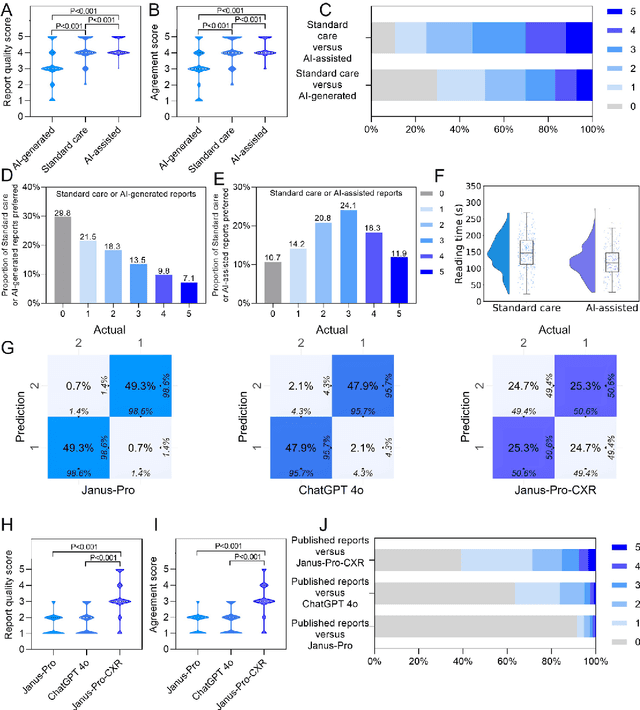
Abstract:A global shortage of radiologists has been exacerbated by the significant volume of chest X-ray workloads, particularly in primary care. Although multimodal large language models show promise, existing evaluations predominantly rely on automated metrics or retrospective analyses, lacking rigorous prospective clinical validation. Janus-Pro-CXR (1B), a chest X-ray interpretation system based on DeepSeek Janus-Pro model, was developed and rigorously validated through a multicenter prospective trial (NCT07117266). Our system outperforms state-of-the-art X-ray report generation models in automated report generation, surpassing even larger-scale models including ChatGPT 4o (200B parameters), while demonstrating reliable detection of six clinically critical radiographic findings. Retrospective evaluation confirms significantly higher report accuracy than Janus-Pro and ChatGPT 4o. In prospective clinical deployment, AI assistance significantly improved report quality scores, reduced interpretation time by 18.3% (P < 0.001), and was preferred by a majority of experts in 54.3% of cases. Through lightweight architecture and domain-specific optimization, Janus-Pro-CXR improves diagnostic reliability and workflow efficiency, particularly in resource-constrained settings. The model architecture and implementation framework will be open-sourced to facilitate the clinical translation of AI-assisted radiology solutions.
Rejoining fragmented ancient bamboo slips with physics-driven deep learning
May 13, 2025



Abstract:Bamboo slips are a crucial medium for recording ancient civilizations in East Asia, and offers invaluable archaeological insights for reconstructing the Silk Road, studying material culture exchanges, and global history. However, many excavated bamboo slips have been fragmented into thousands of irregular pieces, making their rejoining a vital yet challenging step for understanding their content. Here we introduce WisePanda, a physics-driven deep learning framework designed to rejoin fragmented bamboo slips. Based on the physics of fracture and material deterioration, WisePanda automatically generates synthetic training data that captures the physical properties of bamboo fragmentations. This approach enables the training of a matching network without requiring manually paired samples, providing ranked suggestions to facilitate the rejoining process. Compared to the leading curve matching method, WisePanda increases Top-50 matching accuracy from 36\% to 52\%. Archaeologists using WisePanda have experienced substantial efficiency improvements (approximately 20 times faster) when rejoining fragmented bamboo slips. This research demonstrates that incorporating physical principles into deep learning models can significantly enhance their performance, transforming how archaeologists restore and study fragmented artifacts. WisePanda provides a new paradigm for addressing data scarcity in ancient artifact restoration through physics-driven machine learning.
LiftFeat: 3D Geometry-Aware Local Feature Matching
May 06, 2025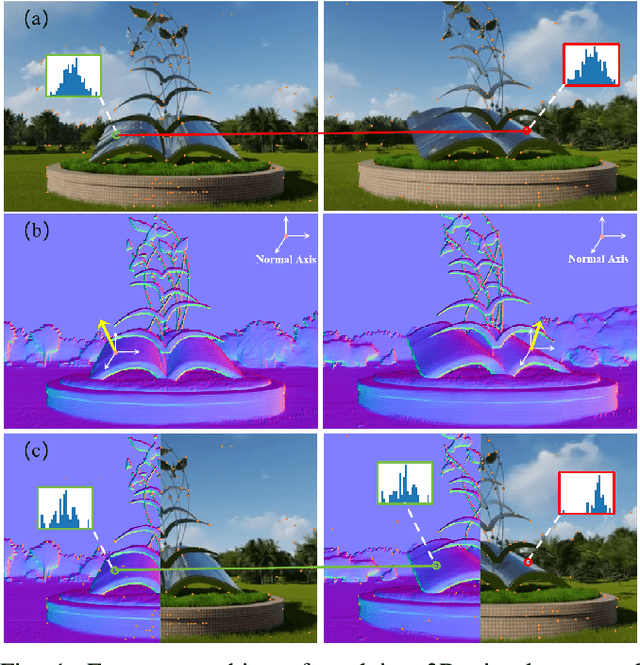
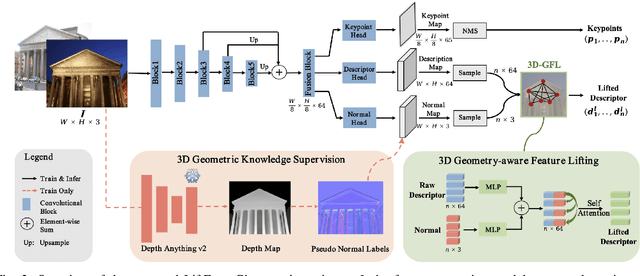
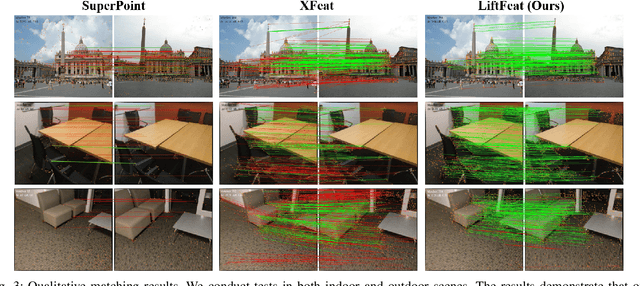
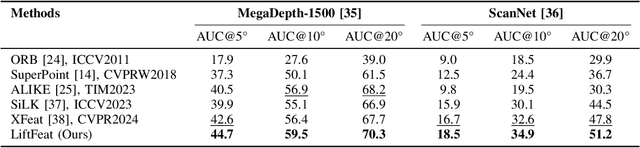
Abstract:Robust and efficient local feature matching plays a crucial role in applications such as SLAM and visual localization for robotics. Despite great progress, it is still very challenging to extract robust and discriminative visual features in scenarios with drastic lighting changes, low texture areas, or repetitive patterns. In this paper, we propose a new lightweight network called \textit{LiftFeat}, which lifts the robustness of raw descriptor by aggregating 3D geometric feature. Specifically, we first adopt a pre-trained monocular depth estimation model to generate pseudo surface normal label, supervising the extraction of 3D geometric feature in terms of predicted surface normal. We then design a 3D geometry-aware feature lifting module to fuse surface normal feature with raw 2D descriptor feature. Integrating such 3D geometric feature enhances the discriminative ability of 2D feature description in extreme conditions. Extensive experimental results on relative pose estimation, homography estimation, and visual localization tasks, demonstrate that our LiftFeat outperforms some lightweight state-of-the-art methods. Code will be released at : https://github.com/lyp-deeplearning/LiftFeat.
Consistent-Point: Consistent Pseudo-Points for Semi-Supervised Crowd Counting and Localization
Mar 16, 2025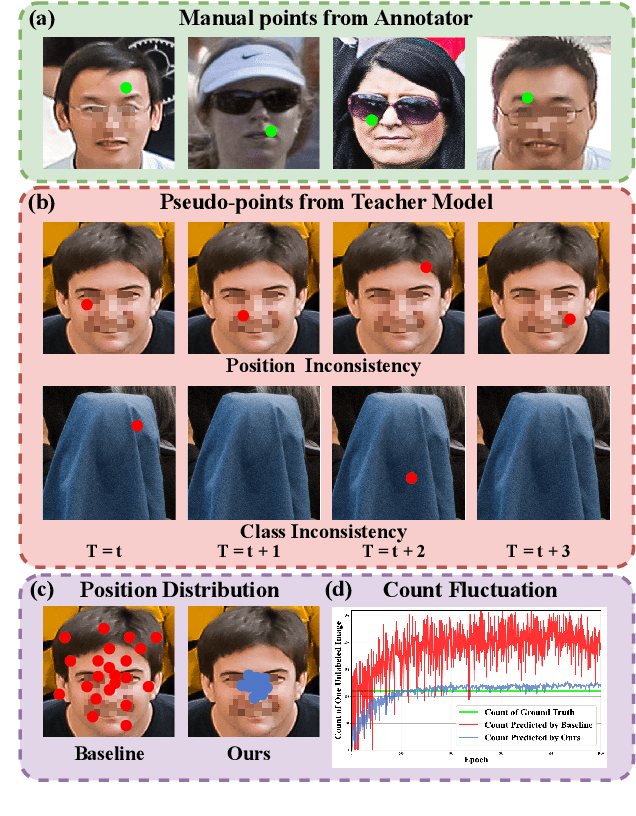
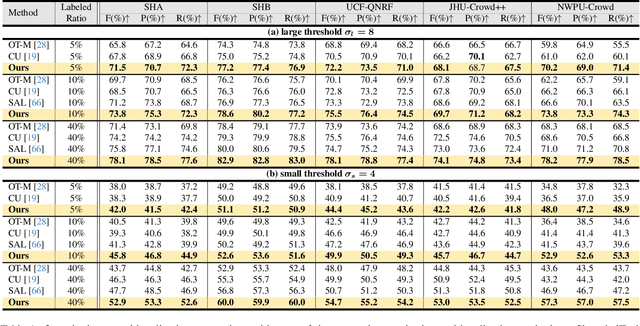
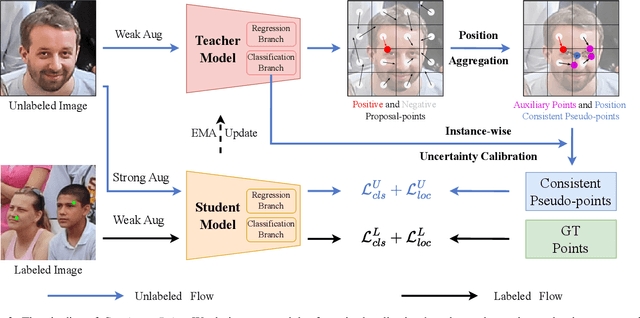
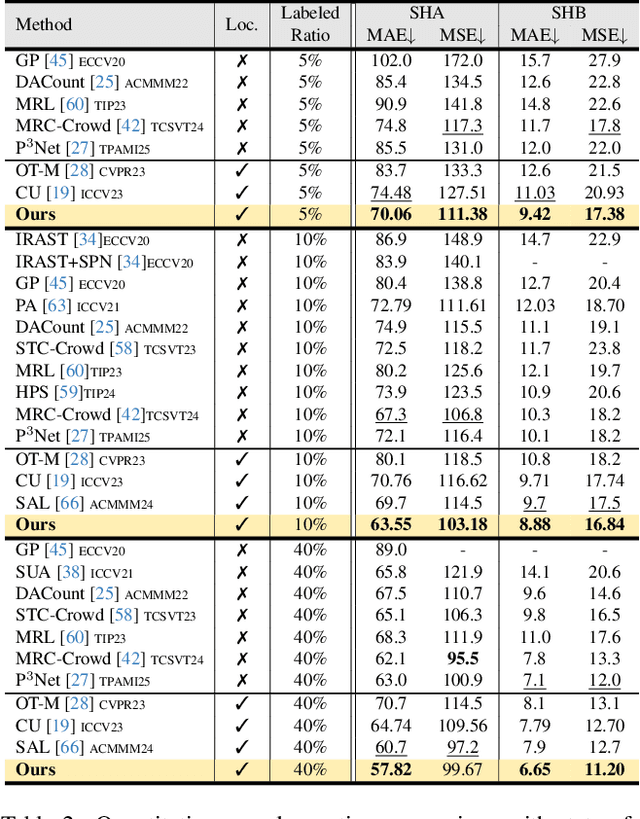
Abstract:Crowd counting and localization are important in applications such as public security and traffic management. Existing methods have achieved impressive results thanks to extensive laborious annotations. This paper propose a novel point-localization-based semi-supervised crowd counting and localization method termed Consistent-Point. We identify and address two inconsistencies of pseudo-points, which have not been adequately explored. To enhance their position consistency, we aggregate the positions of neighboring auxiliary proposal-points. Additionally, an instance-wise uncertainty calibration is proposed to improve the class consistency of pseudo-points. By generating more consistent pseudo-points, Consistent-Point provides more stable supervision to the training process, yielding improved results. Extensive experiments across five widely used datasets and three different labeled ratio settings demonstrate that our method achieves state-of-the-art performance in crowd localization while also attaining impressive crowd counting results. The code will be available.
Pathological Prior-Guided Multiple Instance Learning For Mitigating Catastrophic Forgetting in Breast Cancer Whole Slide Image Classification
Mar 08, 2025Abstract:In histopathology, intelligent diagnosis of Whole Slide Images (WSIs) is essential for automating and objectifying diagnoses, reducing the workload of pathologists. However, diagnostic models often face the challenge of forgetting previously learned data during incremental training on datasets from different sources. To address this issue, we propose a new framework PaGMIL to mitigate catastrophic forgetting in breast cancer WSI classification. Our framework introduces two key components into the common MIL model architecture. First, it leverages microscopic pathological prior to select more accurate and diverse representative patches for MIL. Secondly, it trains separate classification heads for each task and uses macroscopic pathological prior knowledge, treating the thumbnail as a prompt guide (PG) to select the appropriate classification head. We evaluate the continual learning performance of PaGMIL across several public breast cancer datasets. PaGMIL achieves a better balance between the performance of the current task and the retention of previous tasks, outperforming other continual learning methods. Our code will be open-sourced upon acceptance.
MIFNet: Learning Modality-Invariant Features for Generalizable Multimodal Image Matching
Jan 20, 2025



Abstract:Many keypoint detection and description methods have been proposed for image matching or registration. While these methods demonstrate promising performance for single-modality image matching, they often struggle with multimodal data because the descriptors trained on single-modality data tend to lack robustness against the non-linear variations present in multimodal data. Extending such methods to multimodal image matching often requires well-aligned multimodal data to learn modality-invariant descriptors. However, acquiring such data is often costly and impractical in many real-world scenarios. To address this challenge, we propose a modality-invariant feature learning network (MIFNet) to compute modality-invariant features for keypoint descriptions in multimodal image matching using only single-modality training data. Specifically, we propose a novel latent feature aggregation module and a cumulative hybrid aggregation module to enhance the base keypoint descriptors trained on single-modality data by leveraging pre-trained features from Stable Diffusion models. We validate our method with recent keypoint detection and description methods in three multimodal retinal image datasets (CF-FA, CF-OCT, EMA-OCTA) and two remote sensing datasets (Optical-SAR and Optical-NIR). Extensive experiments demonstrate that the proposed MIFNet is able to learn modality-invariant feature for multimodal image matching without accessing the targeted modality and has good zero-shot generalization ability. The source code will be made publicly available.
CellSeg1: Robust Cell Segmentation with One Training Image
Dec 02, 2024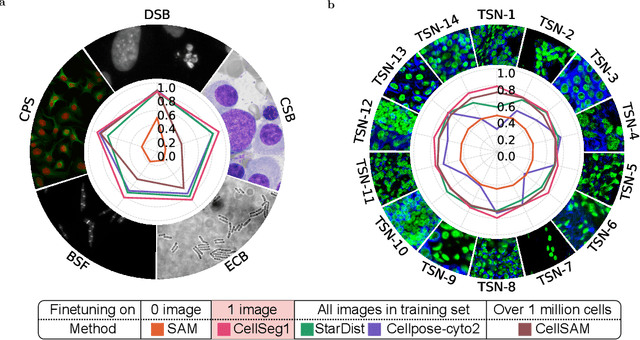
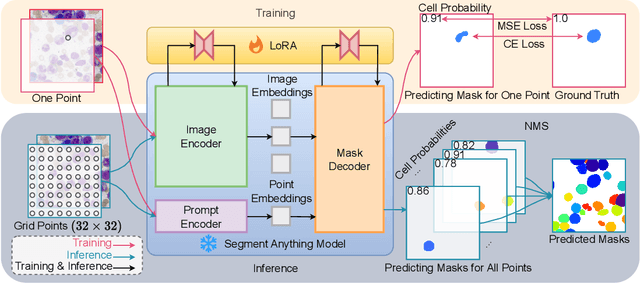
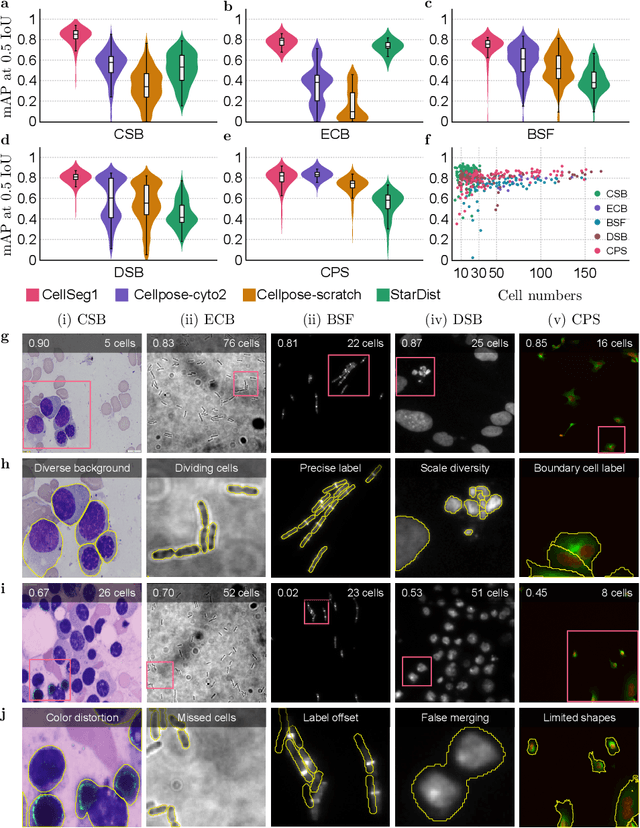
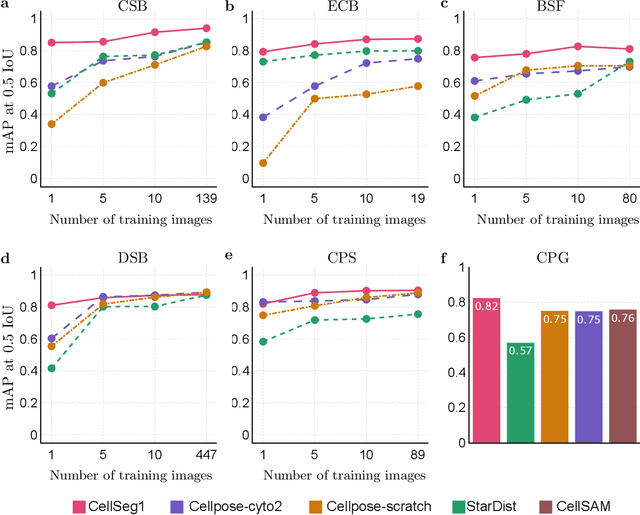
Abstract:Recent trends in cell segmentation have shifted towards universal models to handle diverse cell morphologies and imaging modalities. However, for continuously emerging cell types and imaging techniques, these models still require hundreds or thousands of annotated cells for fine-tuning. We introduce CellSeg1, a practical solution for segmenting cells of arbitrary morphology and modality with a few dozen cell annotations in 1 image. By adopting Low-Rank Adaptation of the Segment Anything Model (SAM), we achieve robust cell segmentation. Tested on 19 diverse cell datasets, CellSeg1 trained on 1 image achieved 0.81 average mAP at 0.5 IoU, performing comparably to existing models trained on over 500 images. It also demonstrated superior generalization in cross-dataset tests on TissueNet. We found that high-quality annotation of a few dozen densely packed cells of varied sizes is key to effective segmentation. CellSeg1 provides an efficient solution for cell segmentation with minimal annotation effort.
Shape Transformation Driven by Active Contour for Class-Imbalanced Semi-Supervised Medical Image Segmentation
Oct 18, 2024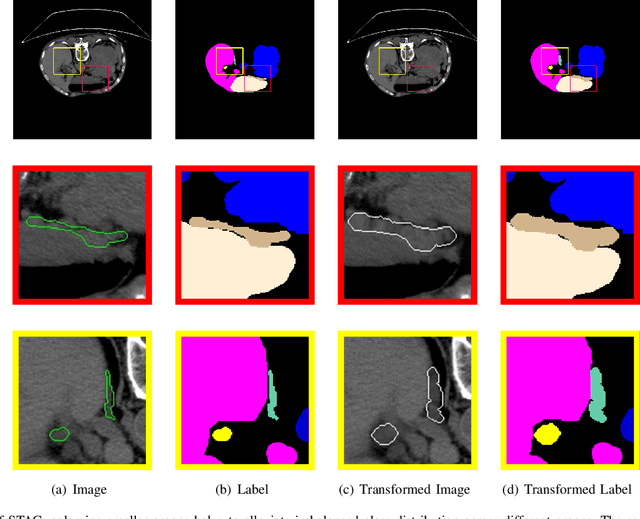
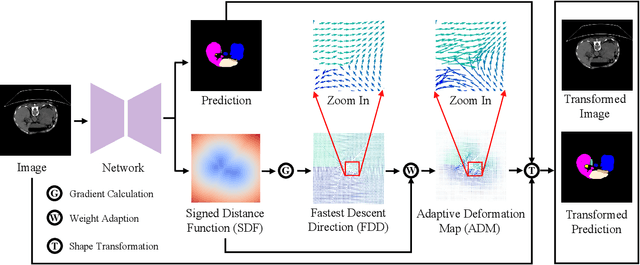
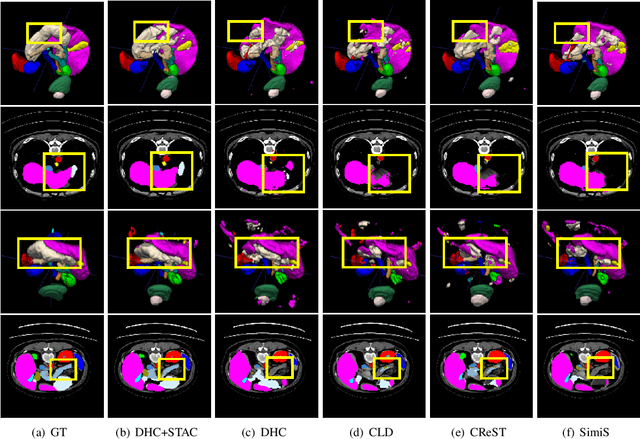
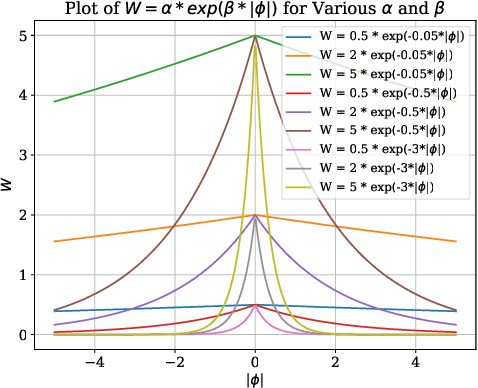
Abstract:Annotating 3D medical images demands expert knowledge and is time-consuming. As a result, semi-supervised learning (SSL) approaches have gained significant interest in 3D medical image segmentation. The significant size differences among various organs in the human body lead to imbalanced class distribution, which is a major challenge in the real-world application of these SSL approaches. To address this issue, we develop a novel Shape Transformation driven by Active Contour (STAC), that enlarges smaller organs to alleviate imbalanced class distribution across different organs. Inspired by curve evolution theory in active contour methods, STAC employs a signed distance function (SDF) as the level set function, to implicitly represent the shape of organs, and deforms voxels in the direction of the steepest descent of SDF (i.e., the normal vector). To ensure that the voxels far from expansion organs remain unchanged, we design an SDF-based weight function to control the degree of deformation for each voxel. We then use STAC as a data-augmentation process during the training stage. Experimental results on two benchmark datasets demonstrate that the proposed method significantly outperforms some state-of-the-art methods. Source code is publicly available at https://github.com/GuGuLL123/STAC.
Shape-intensity knowledge distillation for robust medical image segmentation
Sep 26, 2024Abstract:Many medical image segmentation methods have achieved impressive results. Yet, most existing methods do not take into account the shape-intensity prior information. This may lead to implausible segmentation results, in particular for images of unseen datasets. In this paper, we propose a novel approach to incorporate joint shape-intensity prior information into the segmentation network. Specifically, we first train a segmentation network (regarded as the teacher network) on class-wise averaged training images to extract valuable shape-intensity information, which is then transferred to a student segmentation network with the same network architecture as the teacher via knowledge distillation. In this way, the student network regarded as the final segmentation model can effectively integrate the shape-intensity prior information, yielding more accurate segmentation results. Despite its simplicity, experiments on five medical image segmentation tasks of different modalities demonstrate that the proposed Shape-Intensity Knowledge Distillation (SIKD) consistently improves several baseline models (including recent MaxStyle and SAMed) under intra-dataset evaluation, and significantly improves the cross-dataset generalization ability. The code is available at https://github.com/whdong-whu/SIKD.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge