Yuda Zou
Consistent-Point: Consistent Pseudo-Points for Semi-Supervised Crowd Counting and Localization
Mar 16, 2025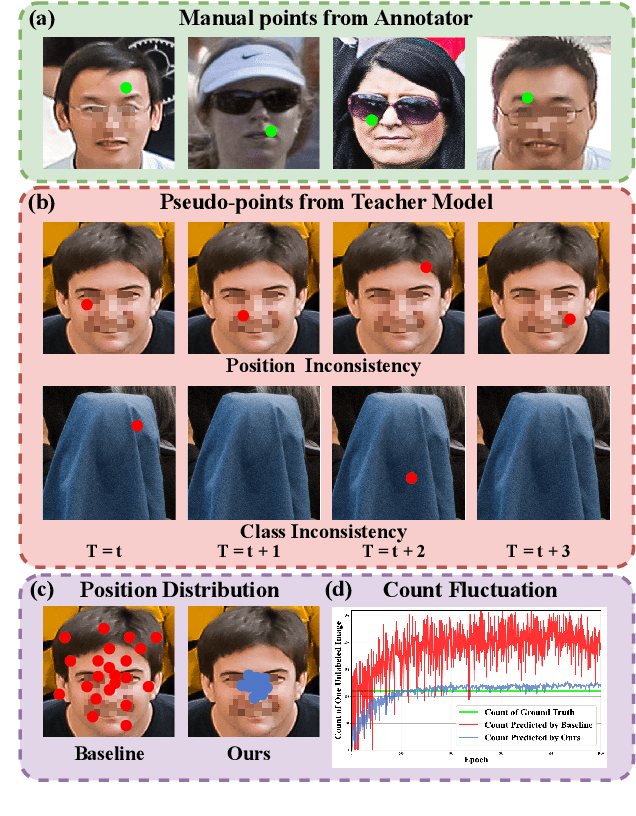
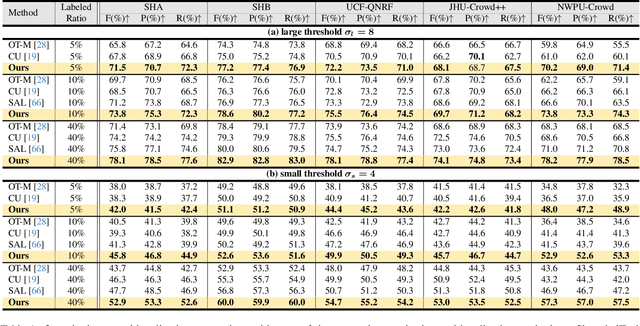
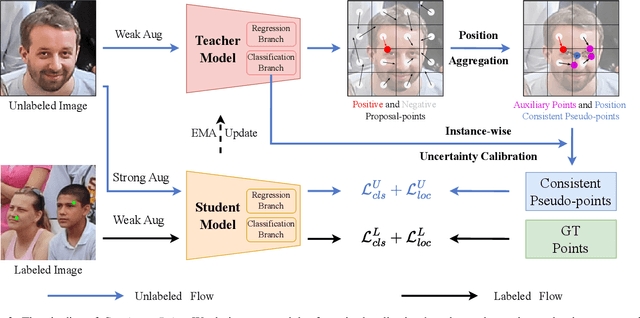
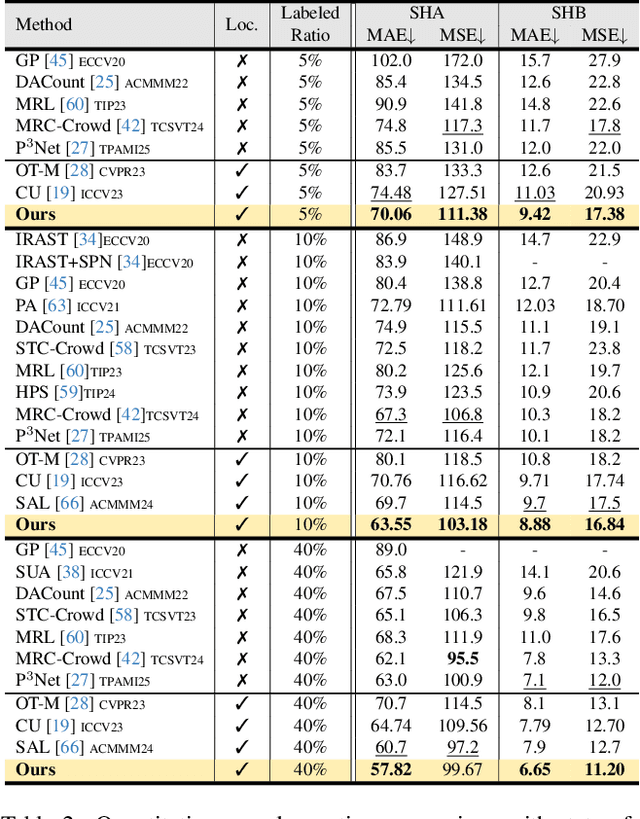
Abstract:Crowd counting and localization are important in applications such as public security and traffic management. Existing methods have achieved impressive results thanks to extensive laborious annotations. This paper propose a novel point-localization-based semi-supervised crowd counting and localization method termed Consistent-Point. We identify and address two inconsistencies of pseudo-points, which have not been adequately explored. To enhance their position consistency, we aggregate the positions of neighboring auxiliary proposal-points. Additionally, an instance-wise uncertainty calibration is proposed to improve the class consistency of pseudo-points. By generating more consistent pseudo-points, Consistent-Point provides more stable supervision to the training process, yielding improved results. Extensive experiments across five widely used datasets and three different labeled ratio settings demonstrate that our method achieves state-of-the-art performance in crowd localization while also attaining impressive crowd counting results. The code will be available.
Dual Structure-Preserving Image Filterings for Semi-supervised Medical Image Segmentation
Dec 12, 2023Abstract:Semi-supervised image segmentation has attracted great attention recently. The key is how to leverage unlabeled images in the training process. Most methods maintain consistent predictions of the unlabeled images under variations (e.g., adding noise/perturbations, or creating alternative versions) in the image and/or model level. In most image-level variation, medical images often have prior structure information, which has not been well explored. In this paper, we propose novel dual structure-preserving image filterings (DSPIF) as the image-level variations for semi-supervised medical image segmentation. Motivated by connected filtering that simplifies image via filtering in structure-aware tree-based image representation, we resort to the dual contrast invariant Max-tree and Min-tree representation. Specifically, we propose a novel connected filtering that removes topologically equivalent nodes (i.e. connected components) having no siblings in the Max/Min-tree. This results in two filtered images preserving topologically critical structure. Applying such dual structure-preserving image filterings in mutual supervision is beneficial for semi-supervised medical image segmentation. Extensive experimental results on three benchmark datasets demonstrate that the proposed method significantly/consistently outperforms some state-of-the-art methods. The source codes will be publicly available.
Noised Autoencoders for Point Annotation Restoration in Object Counting
Dec 12, 2023Abstract:Object counting is a field of growing importance in domains such as security surveillance, urban planning, and biology. The annotation is usually provided in terms of 2D points. However, the complexity of object shapes and subjective of annotators may lead to annotation inconsistency, potentially confusing the model during training. To alleviate this issue, we introduce the Noised Autoencoders (NAE) methodology, which extracts general positional knowledge from all annotations. The method involves adding random offsets to initial point annotations, followed by a UNet to restore them to their original positions. Similar to MAE, NAE faces challenges in restoring non-generic points, necessitating reliance on the most common positions inferred from general knowledge. This reliance forms the cornerstone of our method's effectiveness. Different from existing noise-resistance methods, our approach focus on directly improving initial point annotations. Extensive experiments show that NAE yields more consistent annotations compared to the original ones, steadily enhancing the performance of advanced models trained with these revised annotations. \textbf{Remarkably, the proposed approach helps to set new records in nine datasets}. We will make the NAE codes and refined point annotations available.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge