Jinchi Zhu
LIGM
Rejoining fragmented ancient bamboo slips with physics-driven deep learning
May 13, 2025



Abstract:Bamboo slips are a crucial medium for recording ancient civilizations in East Asia, and offers invaluable archaeological insights for reconstructing the Silk Road, studying material culture exchanges, and global history. However, many excavated bamboo slips have been fragmented into thousands of irregular pieces, making their rejoining a vital yet challenging step for understanding their content. Here we introduce WisePanda, a physics-driven deep learning framework designed to rejoin fragmented bamboo slips. Based on the physics of fracture and material deterioration, WisePanda automatically generates synthetic training data that captures the physical properties of bamboo fragmentations. This approach enables the training of a matching network without requiring manually paired samples, providing ranked suggestions to facilitate the rejoining process. Compared to the leading curve matching method, WisePanda increases Top-50 matching accuracy from 36\% to 52\%. Archaeologists using WisePanda have experienced substantial efficiency improvements (approximately 20 times faster) when rejoining fragmented bamboo slips. This research demonstrates that incorporating physical principles into deep learning models can significantly enhance their performance, transforming how archaeologists restore and study fragmented artifacts. WisePanda provides a new paradigm for addressing data scarcity in ancient artifact restoration through physics-driven machine learning.
LiftFeat: 3D Geometry-Aware Local Feature Matching
May 06, 2025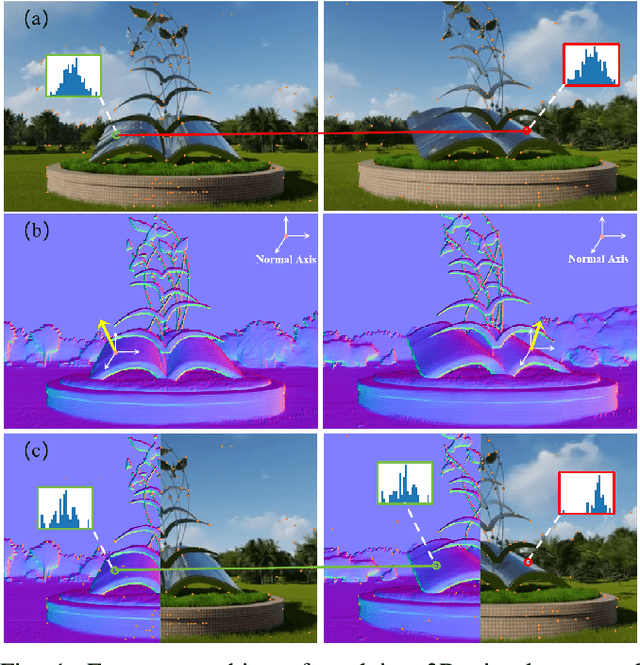
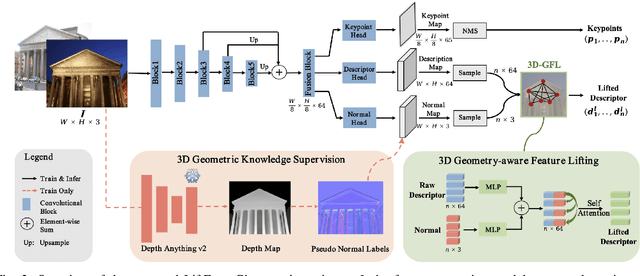
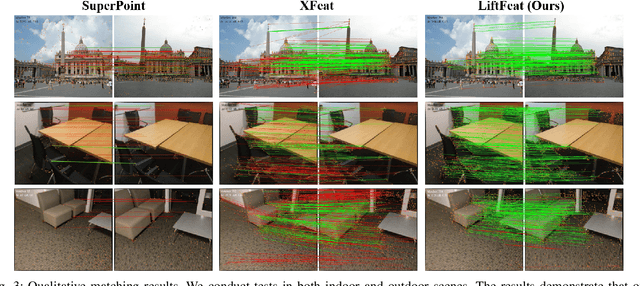
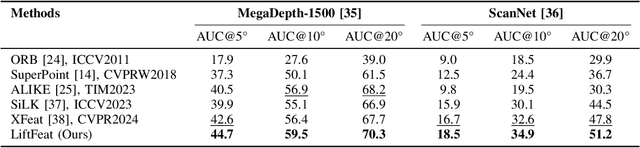
Abstract:Robust and efficient local feature matching plays a crucial role in applications such as SLAM and visual localization for robotics. Despite great progress, it is still very challenging to extract robust and discriminative visual features in scenarios with drastic lighting changes, low texture areas, or repetitive patterns. In this paper, we propose a new lightweight network called \textit{LiftFeat}, which lifts the robustness of raw descriptor by aggregating 3D geometric feature. Specifically, we first adopt a pre-trained monocular depth estimation model to generate pseudo surface normal label, supervising the extraction of 3D geometric feature in terms of predicted surface normal. We then design a 3D geometry-aware feature lifting module to fuse surface normal feature with raw 2D descriptor feature. Integrating such 3D geometric feature enhances the discriminative ability of 2D feature description in extreme conditions. Extensive experimental results on relative pose estimation, homography estimation, and visual localization tasks, demonstrate that our LiftFeat outperforms some lightweight state-of-the-art methods. Code will be released at : https://github.com/lyp-deeplearning/LiftFeat.
Shape Transformation Driven by Active Contour for Class-Imbalanced Semi-Supervised Medical Image Segmentation
Oct 18, 2024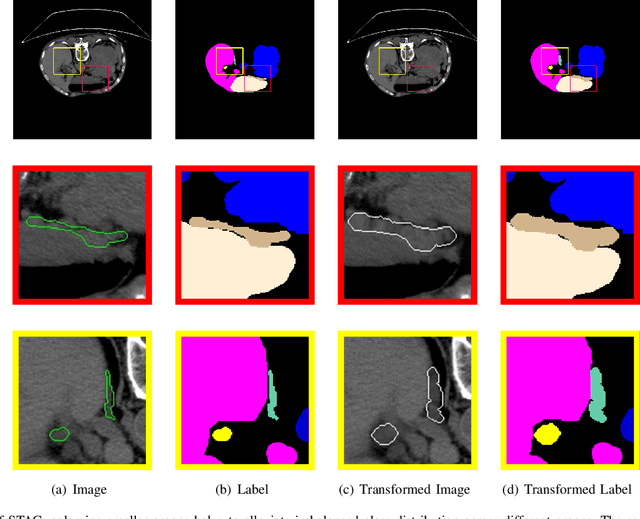
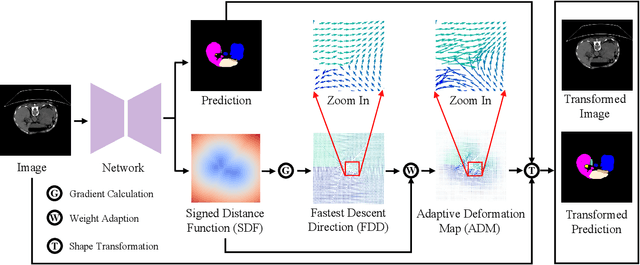
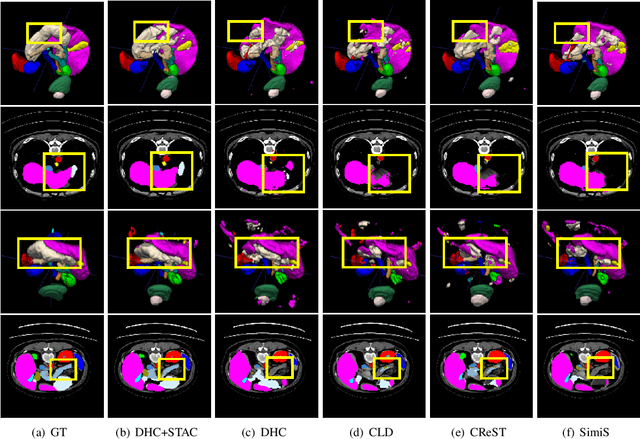
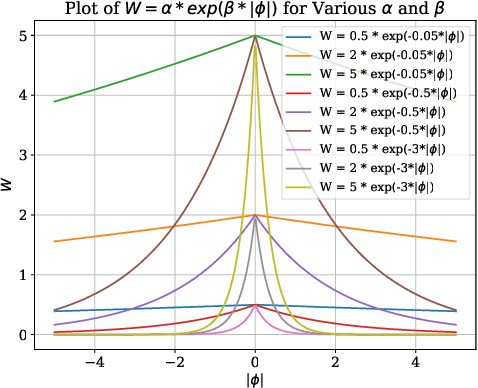
Abstract:Annotating 3D medical images demands expert knowledge and is time-consuming. As a result, semi-supervised learning (SSL) approaches have gained significant interest in 3D medical image segmentation. The significant size differences among various organs in the human body lead to imbalanced class distribution, which is a major challenge in the real-world application of these SSL approaches. To address this issue, we develop a novel Shape Transformation driven by Active Contour (STAC), that enlarges smaller organs to alleviate imbalanced class distribution across different organs. Inspired by curve evolution theory in active contour methods, STAC employs a signed distance function (SDF) as the level set function, to implicitly represent the shape of organs, and deforms voxels in the direction of the steepest descent of SDF (i.e., the normal vector). To ensure that the voxels far from expansion organs remain unchanged, we design an SDF-based weight function to control the degree of deformation for each voxel. We then use STAC as a data-augmentation process during the training stage. Experimental results on two benchmark datasets demonstrate that the proposed method significantly outperforms some state-of-the-art methods. Source code is publicly available at https://github.com/GuGuLL123/STAC.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge