Wenhui Dong
Small-Large Collaboration: Training-efficient Concept Personalization for Large VLM using a Meta Personalized Small VLM
Aug 10, 2025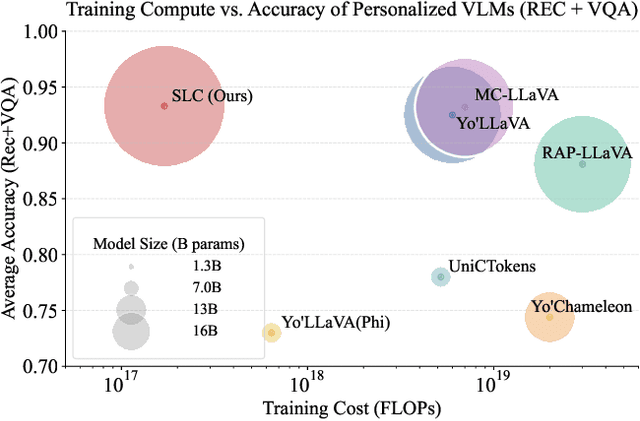
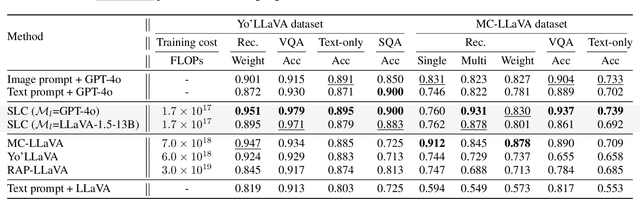
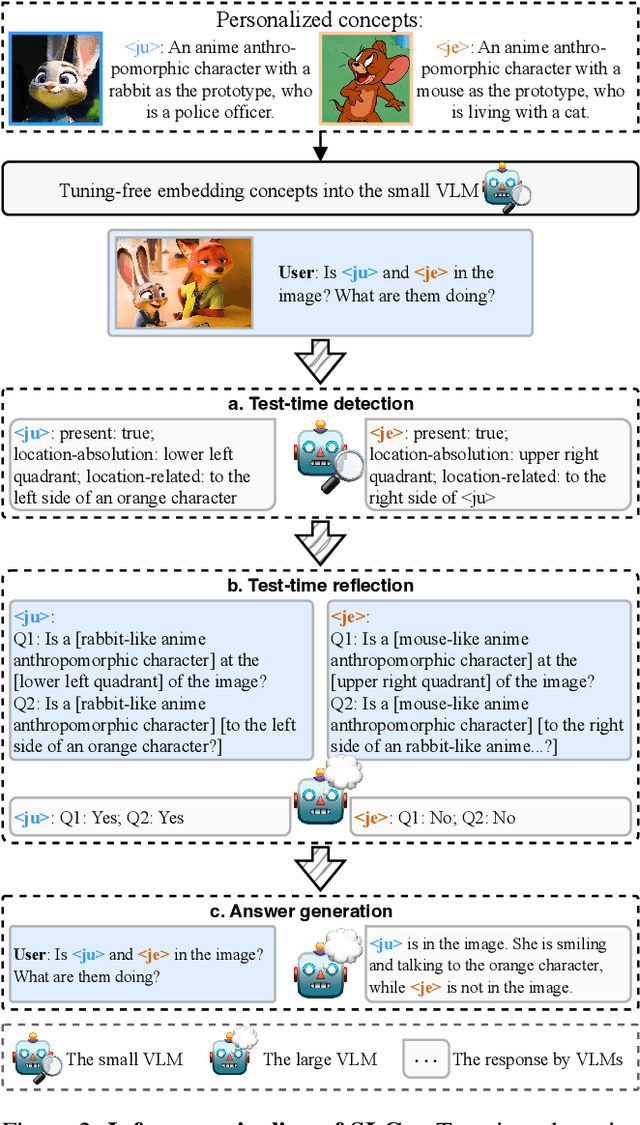
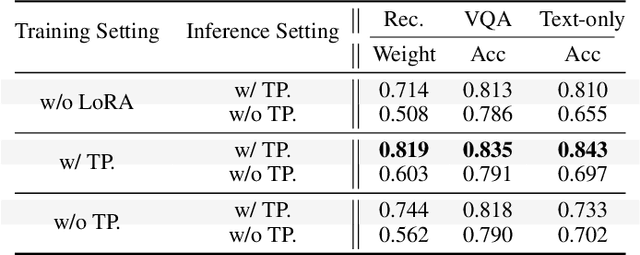
Abstract:Personalizing Vision-Language Models (VLMs) to transform them into daily assistants has emerged as a trending research direction. However, leading companies like OpenAI continue to increase model size and develop complex designs such as the chain of thought (CoT). While large VLMs are proficient in complex multi-modal understanding, their high training costs and limited access via paid APIs restrict direct personalization. Conversely, small VLMs are easily personalized and freely available, but they lack sufficient reasoning capabilities. Inspired by this, we propose a novel collaborative framework named Small-Large Collaboration (SLC) for large VLM personalization, where the small VLM is responsible for generating personalized information, while the large model integrates this personalized information to deliver accurate responses. To effectively incorporate personalized information, we develop a test-time reflection strategy, preventing the potential hallucination of the small VLM. Since SLC only needs to train a meta personalized small VLM for the large VLMs, the overall process is training-efficient. To the best of our knowledge, this is the first training-efficient framework that supports both open-source and closed-source large VLMs, enabling broader real-world personalized applications. We conduct thorough experiments across various benchmarks and large VLMs to demonstrate the effectiveness of the proposed SLC framework. The code will be released at https://github.com/Hhankyangg/SLC.
FiLA-Video: Spatio-Temporal Compression for Fine-Grained Long Video Understanding
Apr 29, 2025Abstract:Recent advancements in video understanding within visual large language models (VLLMs) have led to notable progress. However, the complexity of video data and contextual processing limitations still hinder long-video comprehension. A common approach is video feature compression to reduce token input to large language models, yet many methods either fail to prioritize essential features, leading to redundant inter-frame information, or introduce computationally expensive modules.To address these issues, we propose FiLA(Fine-grained Vision Language Model)-Video, a novel framework that leverages a lightweight dynamic-weight multi-frame fusion strategy, which adaptively integrates multiple frames into a single representation while preserving key video information and reducing computational costs. To enhance frame selection for fusion, we introduce a keyframe selection strategy, effectively identifying informative frames from a larger pool for improved summarization. Additionally, we present a simple yet effective long-video training data generation strategy, boosting model performance without extensive manual annotation. Experimental results demonstrate that FiLA-Video achieves superior efficiency and accuracy in long-video comprehension compared to existing methods.
HyViLM: Enhancing Fine-Grained Recognition with a Hybrid Encoder for Vision-Language Models
Dec 11, 2024



Abstract:Recently, there has been growing interest in the capability of multimodal large language models (MLLMs) to process high-resolution images. A common approach currently involves dynamically cropping the original high-resolution image into smaller sub-images, which are then fed into a vision encoder that was pre-trained on lower-resolution images. However, this cropping approach often truncates objects and connected areas in the original image, causing semantic breaks. To address this limitation, we introduce HyViLM, designed to process images of any resolution while retaining the overall context during encoding. Specifically, we: (i) Design a new visual encoder called Hybrid Encoder that not only encodes individual sub-images but also interacts with detailed global visual features, significantly improving the model's ability to encode high-resolution images. (ii) Propose an optimal feature fusion strategy for the dynamic cropping approach, effectively leveraging information from different layers of the vision encoder. Compared with the state-of-the-art MLLMs under the same setting, our HyViLM outperforms existing MLLMs in nine out of ten tasks. Specifically, HyViLM achieves a 9.6% improvement in performance on the TextVQA task and a 6.9% enhancement on the DocVQA task.
Shape-intensity knowledge distillation for robust medical image segmentation
Sep 26, 2024Abstract:Many medical image segmentation methods have achieved impressive results. Yet, most existing methods do not take into account the shape-intensity prior information. This may lead to implausible segmentation results, in particular for images of unseen datasets. In this paper, we propose a novel approach to incorporate joint shape-intensity prior information into the segmentation network. Specifically, we first train a segmentation network (regarded as the teacher network) on class-wise averaged training images to extract valuable shape-intensity information, which is then transferred to a student segmentation network with the same network architecture as the teacher via knowledge distillation. In this way, the student network regarded as the final segmentation model can effectively integrate the shape-intensity prior information, yielding more accurate segmentation results. Despite its simplicity, experiments on five medical image segmentation tasks of different modalities demonstrate that the proposed Shape-Intensity Knowledge Distillation (SIKD) consistently improves several baseline models (including recent MaxStyle and SAMed) under intra-dataset evaluation, and significantly improves the cross-dataset generalization ability. The code is available at https://github.com/whdong-whu/SIKD.
MoreStyle: Relax Low-frequency Constraint of Fourier-based Image Reconstruction in Generalizable Medical Image Segmentation
Mar 19, 2024



Abstract:The task of single-source domain generalization (SDG) in medical image segmentation is crucial due to frequent domain shifts in clinical image datasets. To address the challenge of poor generalization across different domains, we introduce a Plug-and-Play module for data augmentation called MoreStyle. MoreStyle diversifies image styles by relaxing low-frequency constraints in Fourier space, guiding the image reconstruction network. With the help of adversarial learning, MoreStyle further expands the style range and pinpoints the most intricate style combinations within latent features. To handle significant style variations, we introduce an uncertainty-weighted loss. This loss emphasizes hard-to-classify pixels resulting only from style shifts while mitigating true hard-to-classify pixels in both MoreStyle-generated and original images. Extensive experiments on two widely used benchmarks demonstrate that the proposed MoreStyle effectively helps to achieve good domain generalization ability, and has the potential to further boost the performance of some state-of-the-art SDG methods.
Scale-aware Test-time Click Adaptation for Pulmonary Nodule and Mass Segmentation
Jul 28, 2023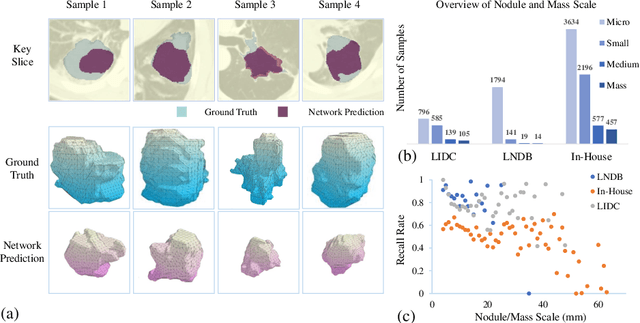
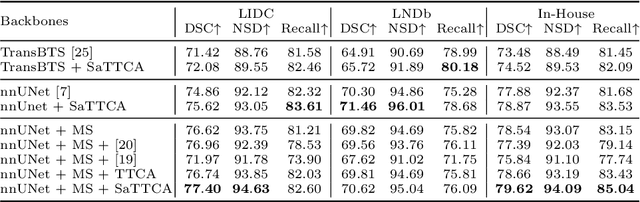
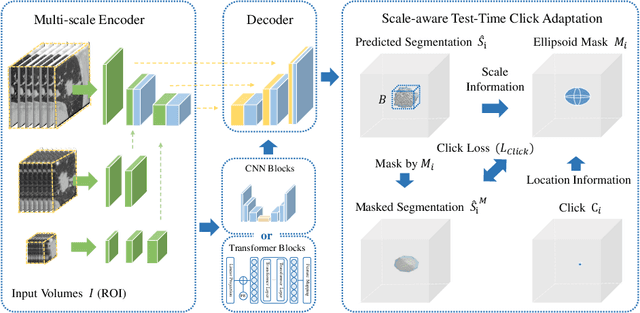
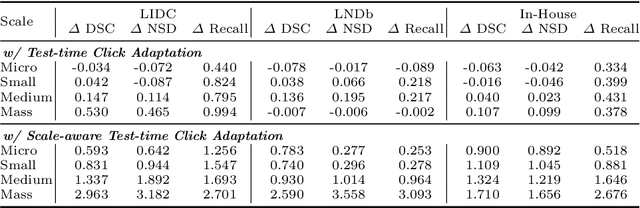
Abstract:Pulmonary nodules and masses are crucial imaging features in lung cancer screening that require careful management in clinical diagnosis. Despite the success of deep learning-based medical image segmentation, the robust performance on various sizes of lesions of nodule and mass is still challenging. In this paper, we propose a multi-scale neural network with scale-aware test-time adaptation to address this challenge. Specifically, we introduce an adaptive Scale-aware Test-time Click Adaptation method based on effortlessly obtainable lesion clicks as test-time cues to enhance segmentation performance, particularly for large lesions. The proposed method can be seamlessly integrated into existing networks. Extensive experiments on both open-source and in-house datasets consistently demonstrate the effectiveness of the proposed method over some CNN and Transformer-based segmentation methods. Our code is available at https://github.com/SplinterLi/SaTTCA
Less is More: Focus Attention for Efficient DETR
Jul 24, 2023Abstract:DETR-like models have significantly boosted the performance of detectors and even outperformed classical convolutional models. However, all tokens are treated equally without discrimination brings a redundant computational burden in the traditional encoder structure. The recent sparsification strategies exploit a subset of informative tokens to reduce attention complexity maintaining performance through the sparse encoder. But these methods tend to rely on unreliable model statistics. Moreover, simply reducing the token population hinders the detection performance to a large extent, limiting the application of these sparse models. We propose Focus-DETR, which focuses attention on more informative tokens for a better trade-off between computation efficiency and model accuracy. Specifically, we reconstruct the encoder with dual attention, which includes a token scoring mechanism that considers both localization and category semantic information of the objects from multi-scale feature maps. We efficiently abandon the background queries and enhance the semantic interaction of the fine-grained object queries based on the scores. Compared with the state-of-the-art sparse DETR-like detectors under the same setting, our Focus-DETR gets comparable complexity while achieving 50.4AP (+2.2) on COCO. The code is available at https://github.com/huawei-noah/noah-research/tree/master/Focus-DETR and https://gitee.com/mindspore/models/tree/master/research/cv/Focus-DETR.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge