David Fleet
TxGemma: Efficient and Agentic LLMs for Therapeutics
Apr 08, 2025Abstract:Therapeutic development is a costly and high-risk endeavor that is often plagued by high failure rates. To address this, we introduce TxGemma, a suite of efficient, generalist large language models (LLMs) capable of therapeutic property prediction as well as interactive reasoning and explainability. Unlike task-specific models, TxGemma synthesizes information from diverse sources, enabling broad application across the therapeutic development pipeline. The suite includes 2B, 9B, and 27B parameter models, fine-tuned from Gemma-2 on a comprehensive dataset of small molecules, proteins, nucleic acids, diseases, and cell lines. Across 66 therapeutic development tasks, TxGemma achieved superior or comparable performance to the state-of-the-art generalist model on 64 (superior on 45), and against state-of-the-art specialist models on 50 (superior on 26). Fine-tuning TxGemma models on therapeutic downstream tasks, such as clinical trial adverse event prediction, requires less training data than fine-tuning base LLMs, making TxGemma suitable for data-limited applications. Beyond these predictive capabilities, TxGemma features conversational models that bridge the gap between general LLMs and specialized property predictors. These allow scientists to interact in natural language, provide mechanistic reasoning for predictions based on molecular structure, and engage in scientific discussions. Building on this, we further introduce Agentic-Tx, a generalist therapeutic agentic system powered by Gemini 2.5 that reasons, acts, manages diverse workflows, and acquires external domain knowledge. Agentic-Tx surpasses prior leading models on the Humanity's Last Exam benchmark (Chemistry & Biology) with 52.3% relative improvement over o3-mini (high) and 26.7% over o3-mini (high) on GPQA (Chemistry) and excels with improvements of 6.3% (ChemBench-Preference) and 2.4% (ChemBench-Mini) over o3-mini (high).
Tx-LLM: A Large Language Model for Therapeutics
Jun 10, 2024



Abstract:Developing therapeutics is a lengthy and expensive process that requires the satisfaction of many different criteria, and AI models capable of expediting the process would be invaluable. However, the majority of current AI approaches address only a narrowly defined set of tasks, often circumscribed within a particular domain. To bridge this gap, we introduce Tx-LLM, a generalist large language model (LLM) fine-tuned from PaLM-2 which encodes knowledge about diverse therapeutic modalities. Tx-LLM is trained using a collection of 709 datasets that target 66 tasks spanning various stages of the drug discovery pipeline. Using a single set of weights, Tx-LLM simultaneously processes a wide variety of chemical or biological entities(small molecules, proteins, nucleic acids, cell lines, diseases) interleaved with free-text, allowing it to predict a broad range of associated properties, achieving competitive with state-of-the-art (SOTA) performance on 43 out of 66 tasks and exceeding SOTA on 22. Among these, Tx-LLM is particularly powerful and exceeds best-in-class performance on average for tasks combining molecular SMILES representations with text such as cell line names or disease names, likely due to context learned during pretraining. We observe evidence of positive transfer between tasks with diverse drug types (e.g.,tasks involving small molecules and tasks involving proteins), and we study the impact of model size, domain finetuning, and prompting strategies on performance. We believe Tx-LLM represents an important step towards LLMs encoding biochemical knowledge and could have a future role as an end-to-end tool across the drug discovery development pipeline.
Advancing Multimodal Medical Capabilities of Gemini
May 06, 2024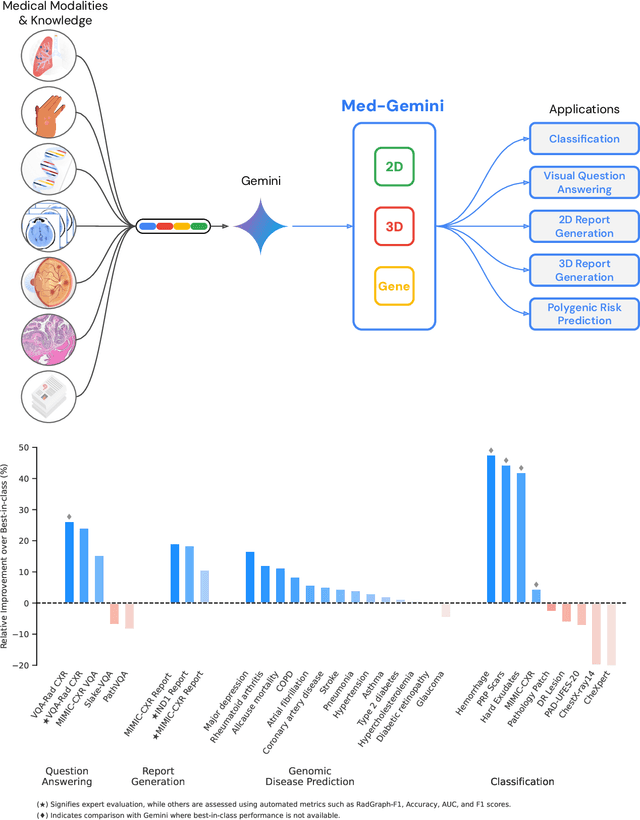
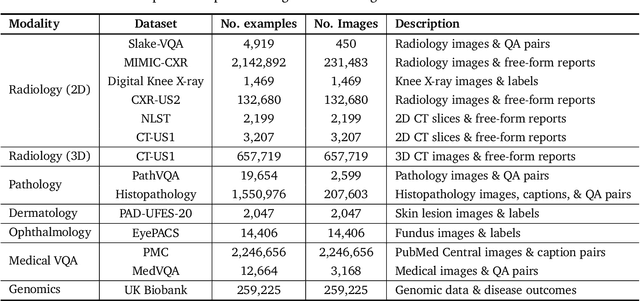

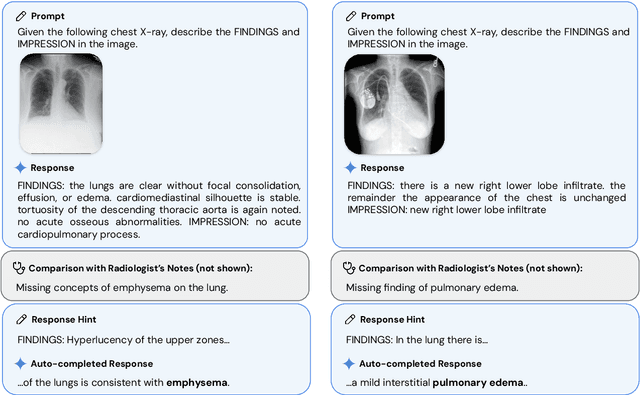
Abstract:Many clinical tasks require an understanding of specialized data, such as medical images and genomics, which is not typically found in general-purpose large multimodal models. Building upon Gemini's multimodal models, we develop several models within the new Med-Gemini family that inherit core capabilities of Gemini and are optimized for medical use via fine-tuning with 2D and 3D radiology, histopathology, ophthalmology, dermatology and genomic data. Med-Gemini-2D sets a new standard for AI-based chest X-ray (CXR) report generation based on expert evaluation, exceeding previous best results across two separate datasets by an absolute margin of 1% and 12%, where 57% and 96% of AI reports on normal cases, and 43% and 65% on abnormal cases, are evaluated as "equivalent or better" than the original radiologists' reports. We demonstrate the first ever large multimodal model-based report generation for 3D computed tomography (CT) volumes using Med-Gemini-3D, with 53% of AI reports considered clinically acceptable, although additional research is needed to meet expert radiologist reporting quality. Beyond report generation, Med-Gemini-2D surpasses the previous best performance in CXR visual question answering (VQA) and performs well in CXR classification and radiology VQA, exceeding SoTA or baselines on 17 of 20 tasks. In histopathology, ophthalmology, and dermatology image classification, Med-Gemini-2D surpasses baselines across 18 out of 20 tasks and approaches task-specific model performance. Beyond imaging, Med-Gemini-Polygenic outperforms the standard linear polygenic risk score-based approach for disease risk prediction and generalizes to genetically correlated diseases for which it has never been trained. Although further development and evaluation are necessary in the safety-critical medical domain, our results highlight the potential of Med-Gemini across a wide range of medical tasks.
Towards Generalist Biomedical AI
Jul 26, 2023



Abstract:Medicine is inherently multimodal, with rich data modalities spanning text, imaging, genomics, and more. Generalist biomedical artificial intelligence (AI) systems that flexibly encode, integrate, and interpret this data at scale can potentially enable impactful applications ranging from scientific discovery to care delivery. To enable the development of these models, we first curate MultiMedBench, a new multimodal biomedical benchmark. MultiMedBench encompasses 14 diverse tasks such as medical question answering, mammography and dermatology image interpretation, radiology report generation and summarization, and genomic variant calling. We then introduce Med-PaLM Multimodal (Med-PaLM M), our proof of concept for a generalist biomedical AI system. Med-PaLM M is a large multimodal generative model that flexibly encodes and interprets biomedical data including clinical language, imaging, and genomics with the same set of model weights. Med-PaLM M reaches performance competitive with or exceeding the state of the art on all MultiMedBench tasks, often surpassing specialist models by a wide margin. We also report examples of zero-shot generalization to novel medical concepts and tasks, positive transfer learning across tasks, and emergent zero-shot medical reasoning. To further probe the capabilities and limitations of Med-PaLM M, we conduct a radiologist evaluation of model-generated (and human) chest X-ray reports and observe encouraging performance across model scales. In a side-by-side ranking on 246 retrospective chest X-rays, clinicians express a pairwise preference for Med-PaLM M reports over those produced by radiologists in up to 40.50% of cases, suggesting potential clinical utility. While considerable work is needed to validate these models in real-world use cases, our results represent a milestone towards the development of generalist biomedical AI systems.
Denoising Diffusion Probabilistic Models for Robust Image Super-Resolution in the Wild
Feb 15, 2023
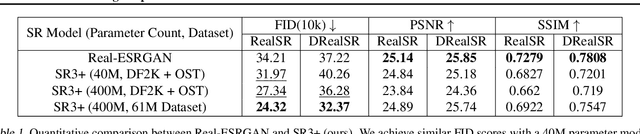
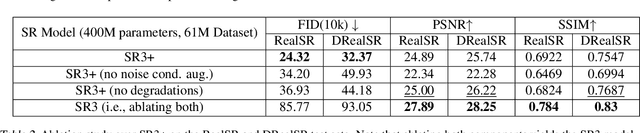
Abstract:Diffusion models have shown promising results on single-image super-resolution and other image- to-image translation tasks. Despite this success, they have not outperformed state-of-the-art GAN models on the more challenging blind super-resolution task, where the input images are out of distribution, with unknown degradations. This paper introduces SR3+, a diffusion-based model for blind super-resolution, establishing a new state-of-the-art. To this end, we advocate self-supervised training with a combination of composite, parameterized degradations for self-supervised training, and noise-conditioing augmentation during training and testing. With these innovations, a large-scale convolutional architecture, and large-scale datasets, SR3+ greatly outperforms SR3. It outperforms Real-ESRGAN when trained on the same data, with a DRealSR FID score of 36.82 vs. 37.22, which further improves to FID of 32.37 with larger models, and further still with larger training sets.
Scalable Adaptive Computation for Iterative Generation
Dec 22, 2022



Abstract:We present the Recurrent Interface Network (RIN), a neural net architecture that allocates computation adaptively to the input according to the distribution of information, allowing it to scale to iterative generation of high-dimensional data. Hidden units of RINs are partitioned into the interface, which is locally connected to inputs, and latents, which are decoupled from inputs and can exchange information globally. The RIN block selectively reads from the interface into latents for high-capacity processing, with incremental updates written back to the interface. Stacking multiple blocks enables effective routing across local and global levels. While routing adds overhead, the cost can be amortized in recurrent computation settings where inputs change gradually while more global context persists, such as iterative generation using diffusion models. To this end, we propose a latent self-conditioning technique that "warm-starts" the latents at each iteration of the generation process. When applied to diffusion models operating directly on pixels, RINs yield state-of-the-art image and video generation without cascades or guidance, while being domain-agnostic and up to 10$\times$ more efficient compared to specialized 2D and 3D U-Nets.
Robust and Efficient Medical Imaging with Self-Supervision
May 19, 2022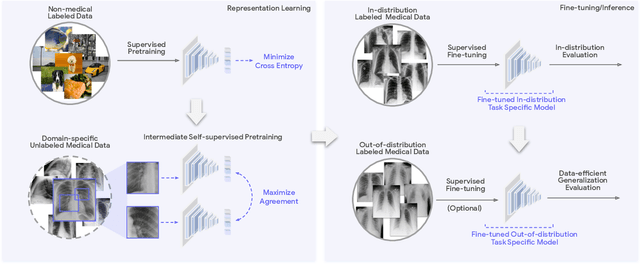
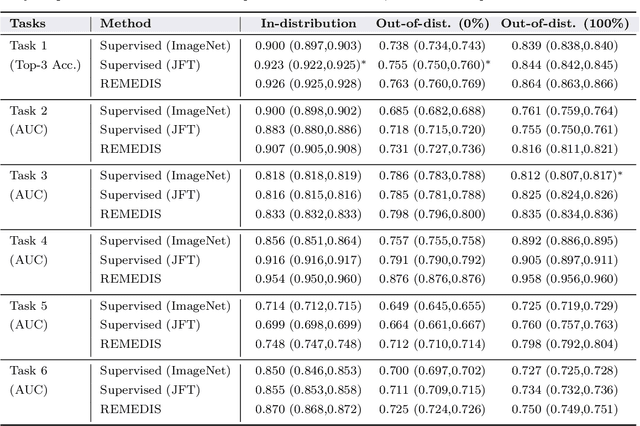
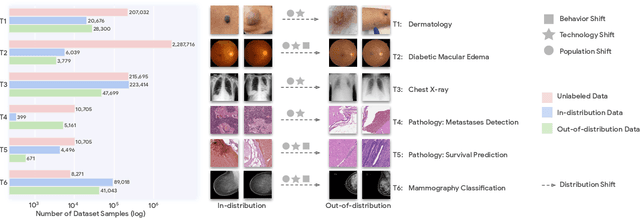
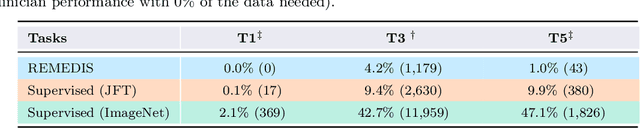
Abstract:Recent progress in Medical Artificial Intelligence (AI) has delivered systems that can reach clinical expert level performance. However, such systems tend to demonstrate sub-optimal "out-of-distribution" performance when evaluated in clinical settings different from the training environment. A common mitigation strategy is to develop separate systems for each clinical setting using site-specific data [1]. However, this quickly becomes impractical as medical data is time-consuming to acquire and expensive to annotate [2]. Thus, the problem of "data-efficient generalization" presents an ongoing difficulty for Medical AI development. Although progress in representation learning shows promise, their benefits have not been rigorously studied, specifically for out-of-distribution settings. To meet these challenges, we present REMEDIS, a unified representation learning strategy to improve robustness and data-efficiency of medical imaging AI. REMEDIS uses a generic combination of large-scale supervised transfer learning with self-supervised learning and requires little task-specific customization. We study a diverse range of medical imaging tasks and simulate three realistic application scenarios using retrospective data. REMEDIS exhibits significantly improved in-distribution performance with up to 11.5% relative improvement in diagnostic accuracy over a strong supervised baseline. More importantly, our strategy leads to strong data-efficient generalization of medical imaging AI, matching strong supervised baselines using between 1% to 33% of retraining data across tasks. These results suggest that REMEDIS can significantly accelerate the life-cycle of medical imaging AI development thereby presenting an important step forward for medical imaging AI to deliver broad impact.
What Makes RAFT Better Than PWC-Net?
Mar 21, 2022
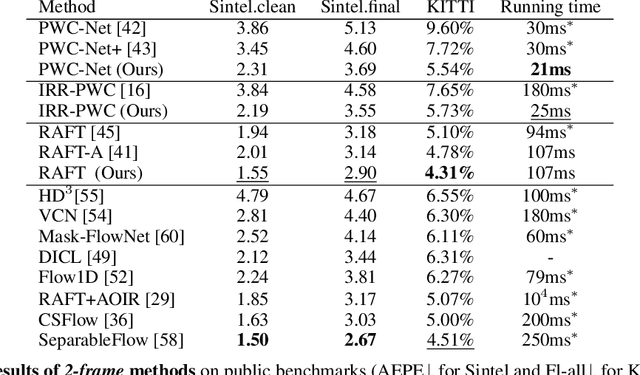

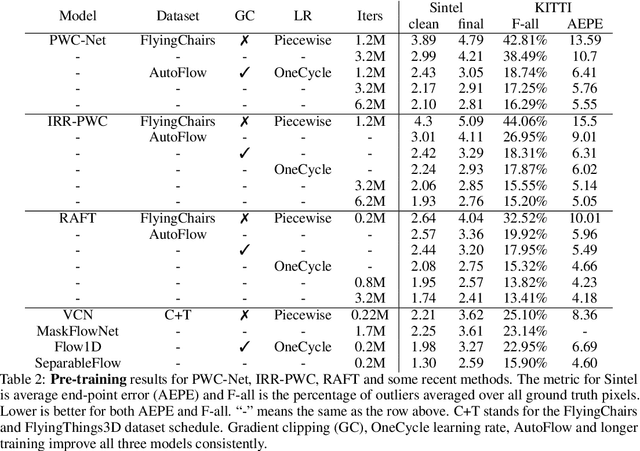
Abstract:How important are training details and datasets to recent optical flow models like RAFT? And do they generalize? To explore these questions, rather than develop a new model, we revisit three prominent models, PWC-Net, IRR-PWC and RAFT, with a common set of modern training techniques and datasets, and observe significant performance gains, demonstrating the importance and generality of these training details. Our newly trained PWC-Net and IRR-PWC models show surprisingly large improvements, up to 30% versus original published results on Sintel and KITTI 2015 benchmarks. They outperform the more recent Flow1D on KITTI 2015 while being 3x faster during inference. Our newly trained RAFT achieves an Fl-all score of 4.31% on KITTI 2015, more accurate than all published optical flow methods at the time of writing. Our results demonstrate the benefits of separating the contributions of models, training techniques and datasets when analyzing performance gains of optical flow methods. Our source code will be publicly available.
Differentiable probabilistic models of scientific imaging with the Fourier slice theorem
Jun 20, 2019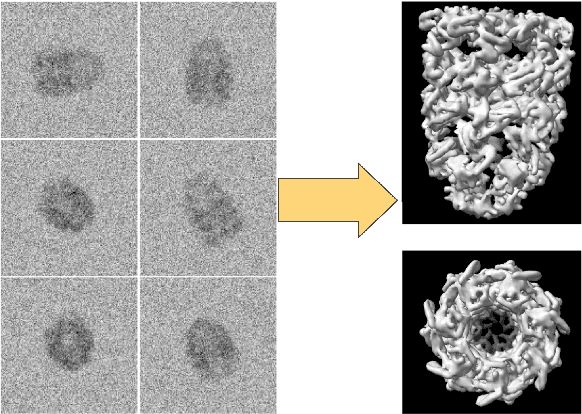

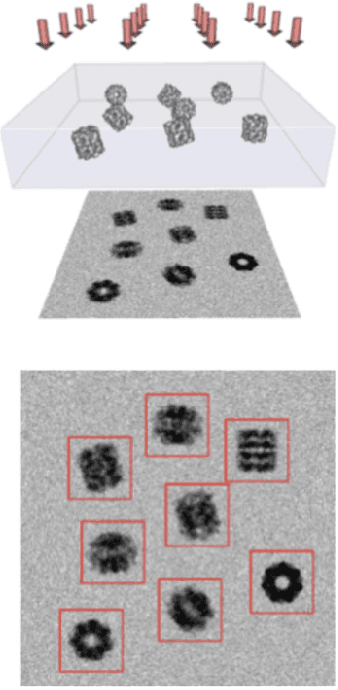
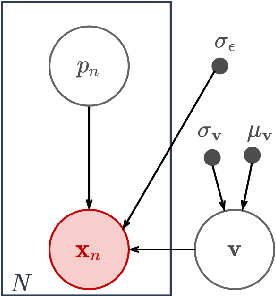
Abstract:Scientific imaging techniques such as optical and electron microscopy and computed tomography (CT) scanning are used to study the 3D structure of an object through 2D observations. These observations are related to the original 3D object through orthogonal integral projections. For common 3D reconstruction algorithms, computational efficiency requires the modeling of the 3D structures to take place in Fourier space by applying the Fourier slice theorem. At present, it is unclear how to differentiate through the projection operator, and hence current learning algorithms can not rely on gradient based methods to optimize 3D structure models. In this paper we show how back-propagation through the projection operator in Fourier space can be achieved. We demonstrate the validity of the approach with experiments on 3D reconstruction of proteins. We further extend our approach to learning probabilistic models of 3D objects. This allows us to predict regions of low sampling rates or estimate noise. A higher sample efficiency can be reached by utilizing the learned uncertainties of the 3D structure as an unsupervised estimate of the model fit. Finally, we demonstrate how the reconstruction algorithm can be extended with an amortized inference scheme on unknown attributes such as object pose. Through empirical studies we show that joint inference of the 3D structure and the object pose becomes more difficult when the ground truth object contains more symmetries. Due to the presence of for instance (approximate) rotational symmetries, the pose estimation can easily get stuck in local optima, inhibiting a fine-grained high-quality estimate of the 3D structure.
TzK: Flow-Based Conditional Generative Model
Mar 14, 2019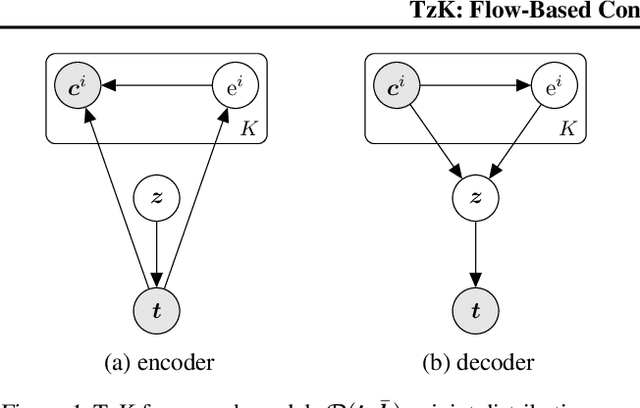
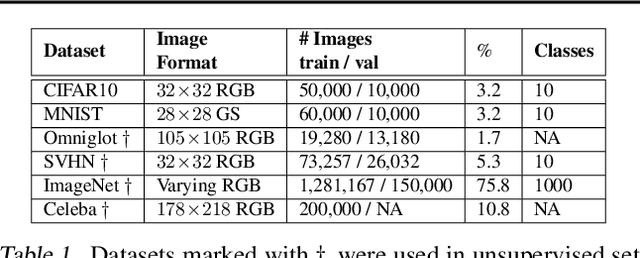


Abstract:We formulate a new class of conditional generative models based on probability flows. Trained with maximum likelihood, it provides efficient inference and sampling from class-conditionals or the joint distribution, and does not require a priori knowledge of the number of classes or the relationships between classes. This allows one to train generative models from multiple, heterogeneous datasets, while retaining strong prior models over subsets of the data (e.g., from a single dataset, class label, or attribute). In this paper, in addition to end-to-end learning, we show how one can learn a single model from multiple datasets with a relatively weak Glow architecture, and then extend it by conditioning on different knowledge types (e.g., a single dataset). This yields log likelihood comparable to state-of-the-art, compelling samples from conditional priors.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge