Micha Livne
cMIM: A Contrastive Mutual Information Framework for Unified Generative and Discriminative Representation Learning
Feb 27, 2025Abstract:Learning representations that are useful for unknown downstream tasks is a fundamental challenge in representation learning. Prominent approaches in this domain include contrastive learning, self-supervised masking, and denoising auto-encoders. In this paper, we introduce a novel method, termed contrastive Mutual Information Machine (cMIM), which aims to enhance the utility of learned representations for downstream tasks. cMIM integrates a new contrastive learning loss with the Mutual Information Machine (MIM) learning framework, a probabilistic auto-encoder that maximizes the mutual information between inputs and latent representations while clustering the latent codes. Despite MIM's potential, initial experiments indicated that the representations learned by MIM were less effective for discriminative downstream tasks compared to state-of-the-art (SOTA) models. The proposed cMIM method directly addresses this limitation. The main contributions of this work are twofold: (1) We propose a novel contrastive extension to MIM for learning discriminative representations which eliminates the need for data augmentation and is robust to variations in the number of negative examples (i.e., batch size). (2) We introduce a generic method for extracting informative embeddings from encoder-decoder models, which significantly improves performance in discriminative downstream tasks without requiring additional training. This method is applicable to any pre-trained encoder-decoder model. By presenting cMIM, we aim to offer a unified generative model that is effective for both generative and discriminative tasks. Our results demonstrate that the learned representations are valuable for downstream tasks while maintaining the generative capabilities of MIM.
BioNeMo Framework: a modular, high-performance library for AI model development in drug discovery
Nov 15, 2024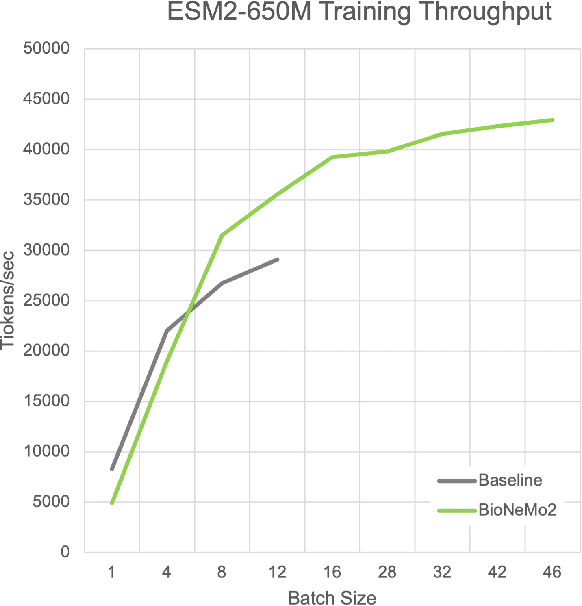
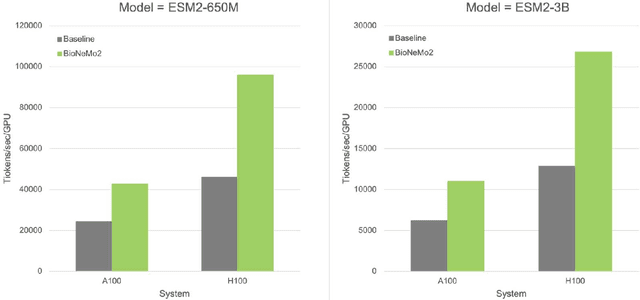
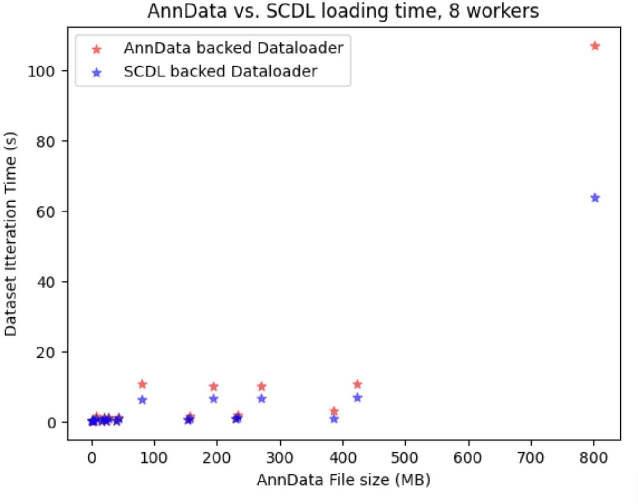
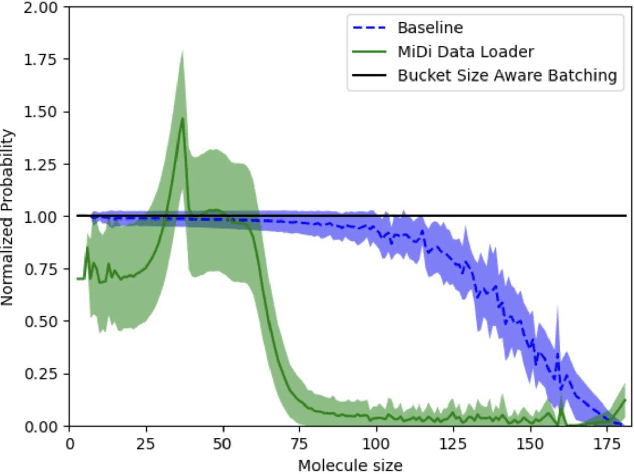
Abstract:Artificial Intelligence models encoding biology and chemistry are opening new routes to high-throughput and high-quality in-silico drug development. However, their training increasingly relies on computational scale, with recent protein language models (pLM) training on hundreds of graphical processing units (GPUs). We introduce the BioNeMo Framework to facilitate the training of computational biology and chemistry AI models across hundreds of GPUs. Its modular design allows the integration of individual components, such as data loaders, into existing workflows and is open to community contributions. We detail technical features of the BioNeMo Framework through use cases such as pLM pre-training and fine-tuning. On 256 NVIDIA A100s, BioNeMo Framework trains a three billion parameter BERT-based pLM on over one trillion tokens in 4.2 days. The BioNeMo Framework is open-source and free for everyone to use.
nach0: Multimodal Natural and Chemical Languages Foundation Model
Nov 21, 2023



Abstract:Large Language Models (LLMs) have substantially driven scientific progress in various domains, and many papers have demonstrated their ability to tackle complex problems with creative solutions. Our paper introduces a new foundation model, nach0, capable of solving various chemical and biological tasks: biomedical question answering, named entity recognition, molecular generation, molecular synthesis, attributes prediction, and others. nach0 is a multi-domain and multi-task encoder-decoder LLM pre-trained on unlabeled text from scientific literature, patents, and molecule strings to incorporate a range of chemical and linguistic knowledge. We employed instruction tuning, where specific task-related instructions are utilized to fine-tune nach0 for the final set of tasks. To train nach0 effectively, we leverage the NeMo framework, enabling efficient parallel optimization of both base and large model versions. Extensive experiments demonstrate that our model outperforms state-of-the-art baselines on single-domain and cross-domain tasks. Furthermore, it can generate high-quality outputs in molecular and textual formats, showcasing its effectiveness in multi-domain setups.
Improving Small Molecule Generation using Mutual Information Machine
Aug 18, 2022
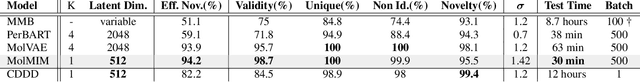
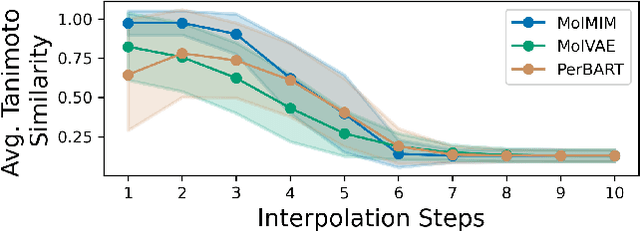
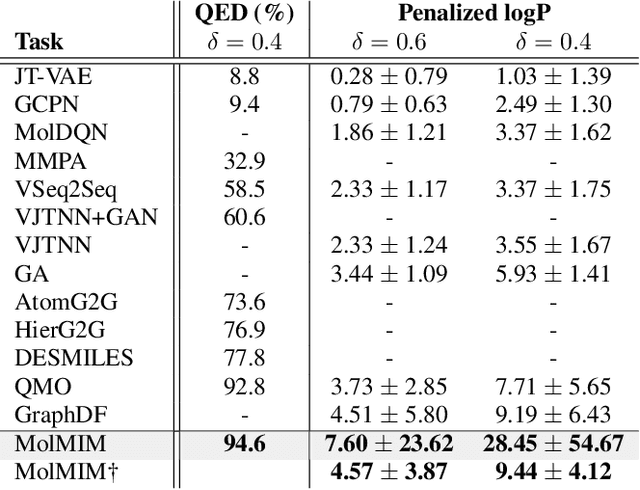
Abstract:We address the task of controlled generation of small molecules, which entails finding novel molecules with desired properties under certain constraints (e.g., similarity to a reference molecule). Here we introduce MolMIM, a probabilistic auto-encoder for small molecule drug discovery that learns an informative and clustered latent space. MolMIM is trained with Mutual Information Machine (MIM) learning, and provides a fixed length representation of variable length SMILES strings. Since encoder-decoder models can learn representations with ``holes'' of invalid samples, here we propose a novel extension to the training procedure which promotes a dense latent space, and allows the model to sample valid molecules from random perturbations of latent codes. We provide a thorough comparison of MolMIM to several variable-size and fixed-size encoder-decoder models, demonstrating MolMIM's superior generation as measured in terms of validity, uniqueness, and novelty. We then utilize CMA-ES, a naive black-box and gradient free search algorithm, over MolMIM's latent space for the task of property guided molecule optimization. We achieve state-of-the-art results in several constrained single property optimization tasks as well as in the challenging task of multi-objective optimization, improving over previous success rate SOTA by more than 5\% . We attribute the strong results to MolMIM's latent representation which clusters similar molecules in the latent space, whereas CMA-ES is often used as a baseline optimization method. We also demonstrate MolMIM to be favourable in a compute limited regime, making it an attractive model for such cases.
SentenceMIM: A Latent Variable Language Model
Mar 06, 2020



Abstract:We introduce sentenceMIM, a probabilistic auto-encoder for language modelling, trained with Mutual Information Machine (MIM) learning. Previous attempts to learn variational auto-encoders for language data have had mixed success, with empirical performance well below state-of-the-art auto-regressive models, a key barrier being the occurrence of posterior collapse with VAEs. The recently proposed MIM framework encourages high mutual information between observations and latent variables, and is more robust against posterior collapse. This paper formulates a MIM model for text data, along with a corresponding learning algorithm. We demonstrate excellent perplexity (PPL) results on several datasets, and show that the framework learns a rich latent space, allowing for interpolation between sentences of different lengths with a fixed-dimensional latent representation. We also demonstrate the versatility of sentenceMIM by utilizing a trained model for question-answering, a transfer learning task, without fine-tuning. To the best of our knowledge, this is the first latent variable model (LVM) for text modelling that achieves competitive performance with non-LVM models.
MIM: Mutual Information Machine
Oct 14, 2019



Abstract:We introduce the Mutual Information Machine (MIM), an autoencoder model for learning joint distributions over observations and latent states. The model formulation reflects two key design principles: 1) symmetry, to encourage the encoder and decoder to learn consistent factorizations of the same underlying distribution; and 2) mutual information, to encourage the learning of useful representations for downstream tasks. The objective comprises the Jensen-Shannon divergence between the encoding and decoding joint distributions, plus a mutual information term. We show that this objective can be bounded by a tractable cross-entropy loss between the true model and a parameterized approximation, and relate this to maximum likelihood estimation and variational autoencoders. Experiments show that MIM is capable of learning a latent representation with high mutual information, and good unsupervised clustering, while providing data log likelihood comparable to VAE (with a sufficiently expressive architecture).
High Mutual Information in Representation Learning with Symmetric Variational Inference
Oct 04, 2019



Abstract:We introduce the Mutual Information Machine (MIM), a novel formulation of representation learning, using a joint distribution over the observations and latent state in an encoder/decoder framework. Our key principles are symmetry and mutual information, where symmetry encourages the encoder and decoder to learn different factorizations of the same underlying distribution, and mutual information, to encourage the learning of useful representations for downstream tasks. Our starting point is the symmetric Jensen-Shannon divergence between the encoding and decoding joint distributions, plus a mutual information encouraging regularizer. We show that this can be bounded by a tractable cross entropy loss function between the true model and a parameterized approximation, and relate this to the maximum likelihood framework. We also relate MIM to variational autoencoders (VAEs) and demonstrate that MIM is capable of learning symmetric factorizations, with high mutual information that avoids posterior collapse.
TzK: Flow-Based Conditional Generative Model
Mar 14, 2019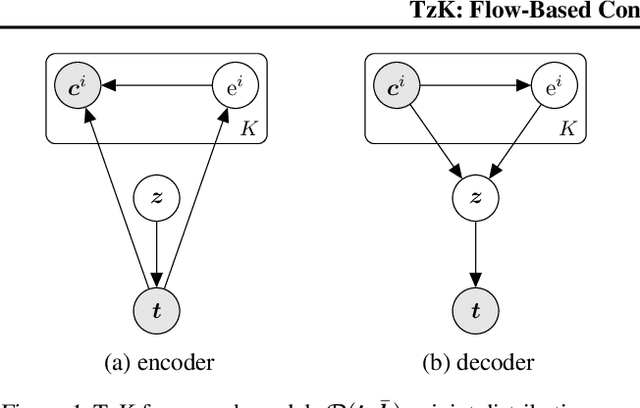


Abstract:We formulate a new class of conditional generative models based on probability flows. Trained with maximum likelihood, it provides efficient inference and sampling from class-conditionals or the joint distribution, and does not require a priori knowledge of the number of classes or the relationships between classes. This allows one to train generative models from multiple, heterogeneous datasets, while retaining strong prior models over subsets of the data (e.g., from a single dataset, class label, or attribute). In this paper, in addition to end-to-end learning, we show how one can learn a single model from multiple datasets with a relatively weak Glow architecture, and then extend it by conditioning on different knowledge types (e.g., a single dataset). This yields log likelihood comparable to state-of-the-art, compelling samples from conditional priors.
Walking on Thin Air: Environment-Free Physics-based Markerless Motion Capture
Dec 04, 2018



Abstract:We propose a generative approach to physics-based motion capture. Unlike prior attempts to incorporate physics into tracking that assume the subject and scene geometry are calibrated and known a priori, our approach is automatic and online. This distinction is important since calibration of the environment is often difficult, especially for motions with props, uneven surfaces, or outdoor scenes. The use of physics in this context provides a natural framework to reason about contact and the plausibility of recovered motions. We propose a fast data-driven parametric body model, based on linear-blend skinning, which decouples deformations due to pose, anthropometrics and body shape. Pose (and shape) parameters are estimated using robust ICP optimization with physics-based dynamic priors that incorporate contact. Contact is estimated from torque trajectories and predictions of which contact points were active. To our knowledge, this is the first approach to take physics into account without explicit {\em a priori} knowledge of the environment or body dimensions. We demonstrate effective tracking from a noisy single depth camera, improving on state-of-the-art results quantitatively and producing better qualitative results, reducing visual artifacts like foot-skate and jitter.
TzK Flow - Conditional Generative Model
Nov 30, 2018



Abstract:We introduce TzK (pronounced "task"), a conditional probability flow-based model that exploits attributes (e.g., style, class membership, or other side information) in order to learn tight conditional prior around manifolds of the target observations. The model is trained via approximated ML, and offers efficient approximation of arbitrary data sample distributions (similar to GAN and flow-based ML), and stable training (similar to VAE and ML), while avoiding variational approximations. TzK exploits meta-data to facilitate a bottleneck, similar to autoencoders, thereby producing a low-dimensional representation. Unlike autoencoders, the bottleneck does not limit model expressiveness, similar to flow-based ML. Supervised, unsupervised, and semi-supervised learning are supported by replacing missing observations with samples from learned priors. We demonstrate TzK by training jointly on MNIST and Omniglot datasets with minimal preprocessing, and weak supervision, with results comparable to state-of-the-art.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge