Daniil Polykovskiy
nach0-pc: Multi-task Language Model with Molecular Point Cloud Encoder
Oct 11, 2024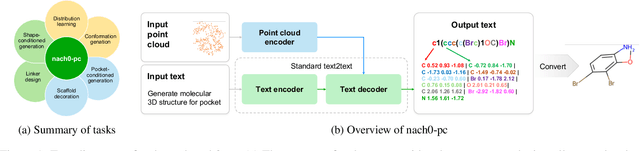
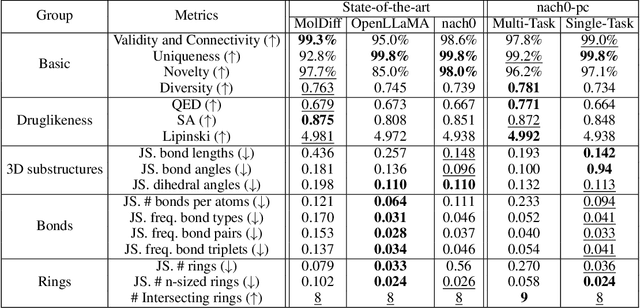
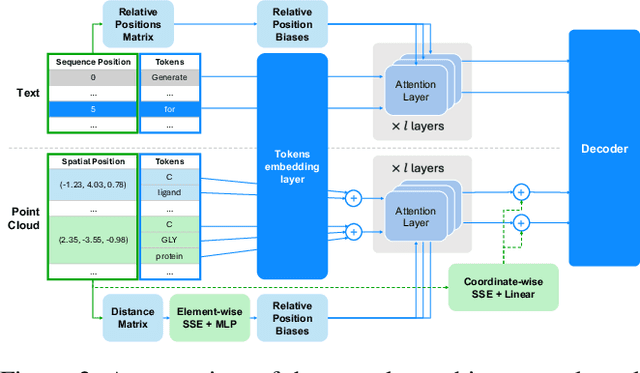
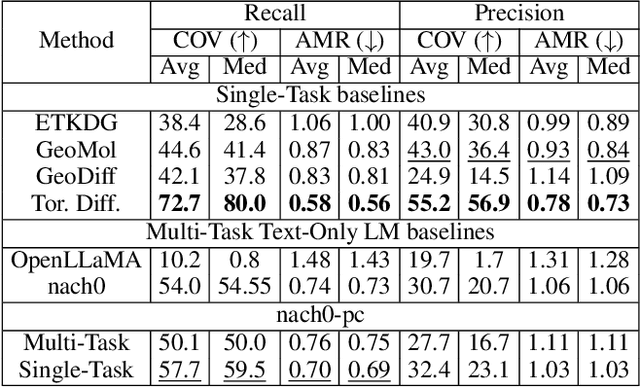
Abstract:Recent advancements have integrated Language Models (LMs) into a drug discovery pipeline. However, existing models mostly work with SMILES and SELFIES chemical string representations, which lack spatial features vital for drug discovery. Additionally, attempts to translate chemical 3D structures into text format encounter issues such as excessive length and insufficient atom connectivity information. To address these issues, we introduce nach0-pc, a model combining domain-specific encoder and textual representation to handle spatial arrangement of atoms effectively. Our approach utilizes a molecular point cloud encoder for concise and order-invariant structure representation. We introduce a novel pre-training scheme for molecular point clouds to distillate the knowledge from spatial molecular structures datasets. After fine-tuning within both single-task and multi-task frameworks, nach0-pc demonstrates performance comparable with other diffusion models in terms of generated samples quality across several established spatial molecular generation tasks. Notably, our model is a multi-task approach, in contrast to diffusion models being limited to single tasks. Additionally, it is capable of processing point cloud-related data, which language models are not capable of handling due to memory limitations. These lead to our model having reduced training and inference time while maintaining on par performance.
BindGPT: A Scalable Framework for 3D Molecular Design via Language Modeling and Reinforcement Learning
Jun 06, 2024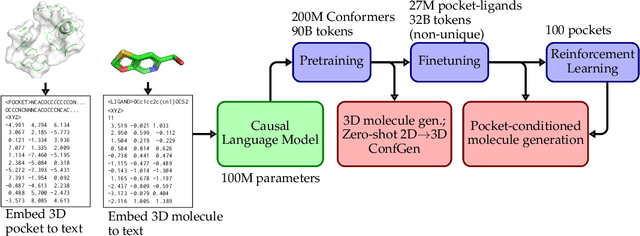
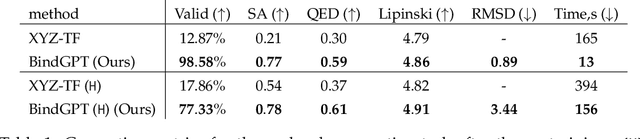
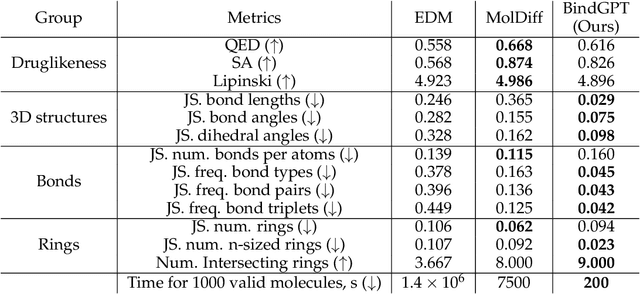
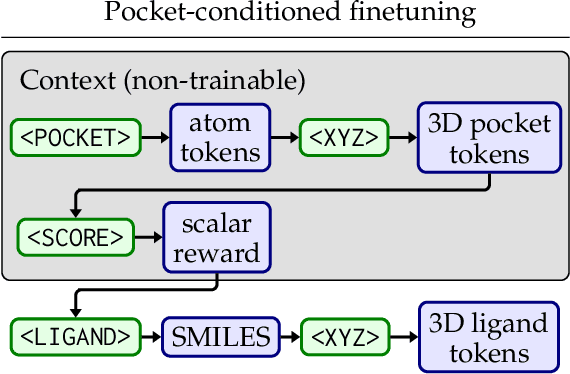
Abstract:Generating novel active molecules for a given protein is an extremely challenging task for generative models that requires an understanding of the complex physical interactions between the molecule and its environment. In this paper, we present a novel generative model, BindGPT which uses a conceptually simple but powerful approach to create 3D molecules within the protein's binding site. Our model produces molecular graphs and conformations jointly, eliminating the need for an extra graph reconstruction step. We pretrain BindGPT on a large-scale dataset and fine-tune it with reinforcement learning using scores from external simulation software. We demonstrate how a single pretrained language model can serve at the same time as a 3D molecular generative model, conformer generator conditioned on the molecular graph, and a pocket-conditioned 3D molecule generator. Notably, the model does not make any representational equivariance assumptions about the domain of generation. We show how such simple conceptual approach combined with pretraining and scaling can perform on par or better than the current best specialized diffusion models, language models, and graph neural networks while being two orders of magnitude cheaper to sample.
nach0: Multimodal Natural and Chemical Languages Foundation Model
Nov 21, 2023



Abstract:Large Language Models (LLMs) have substantially driven scientific progress in various domains, and many papers have demonstrated their ability to tackle complex problems with creative solutions. Our paper introduces a new foundation model, nach0, capable of solving various chemical and biological tasks: biomedical question answering, named entity recognition, molecular generation, molecular synthesis, attributes prediction, and others. nach0 is a multi-domain and multi-task encoder-decoder LLM pre-trained on unlabeled text from scientific literature, patents, and molecule strings to incorporate a range of chemical and linguistic knowledge. We employed instruction tuning, where specific task-related instructions are utilized to fine-tune nach0 for the final set of tasks. To train nach0 effectively, we leverage the NeMo framework, enabling efficient parallel optimization of both base and large model versions. Extensive experiments demonstrate that our model outperforms state-of-the-art baselines on single-domain and cross-domain tasks. Furthermore, it can generate high-quality outputs in molecular and textual formats, showcasing its effectiveness in multi-domain setups.
Chemistry42: An AI-based platform for de novo molecular design
Jan 22, 2021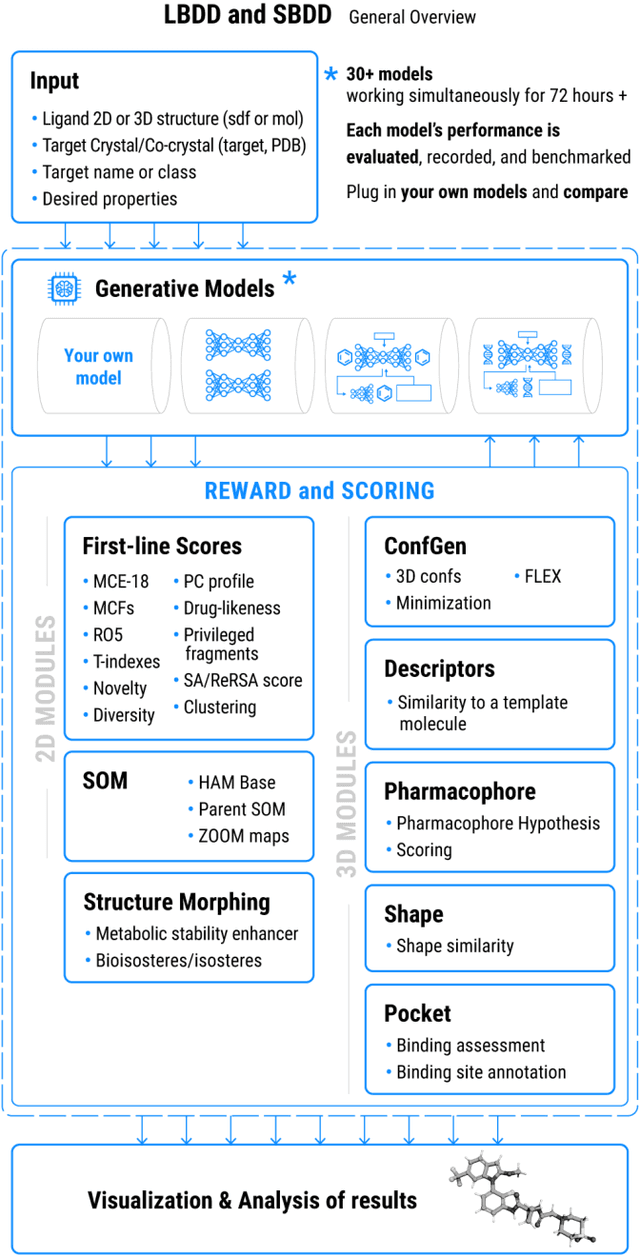
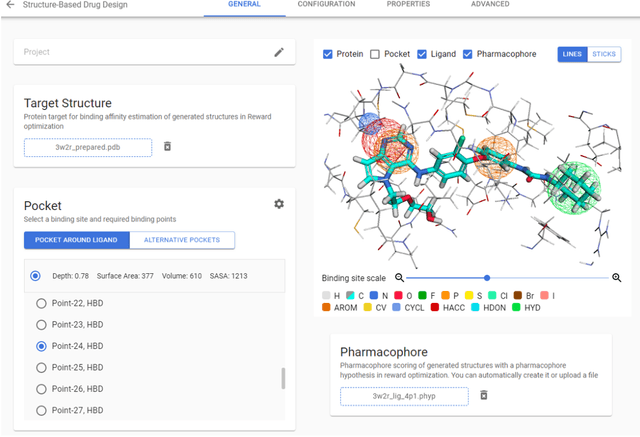
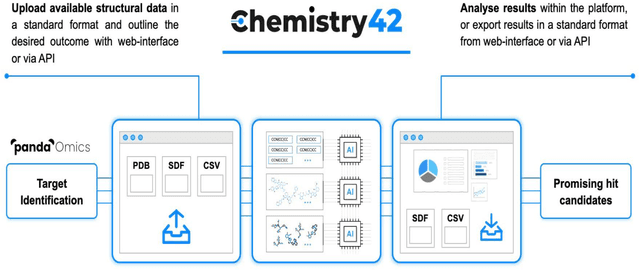
Abstract:Chemistry42 is a software platform for de novo small molecule design that integrates Artificial Intelligence (AI) techniques with computational and medicinal chemistry methods. Chemistry42 is unique in its ability to generate novel molecular structures with predefined properties validated through in vitro and in vivo studies. Chemistry42 is a core component of Insilico Medicine Pharma.ai drug discovery suite that also includes target discovery and multi-omics data analysis (PandaOmics) and clinical trial outcomes predictions (InClinico).
Deterministic Decoding for Discrete Data in Variational Autoencoders
Mar 04, 2020
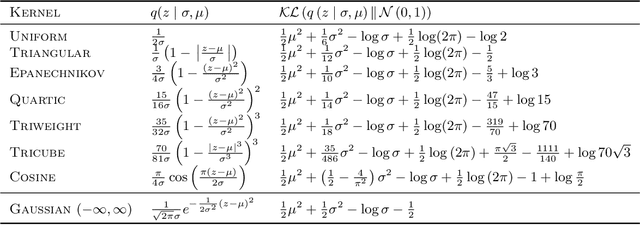

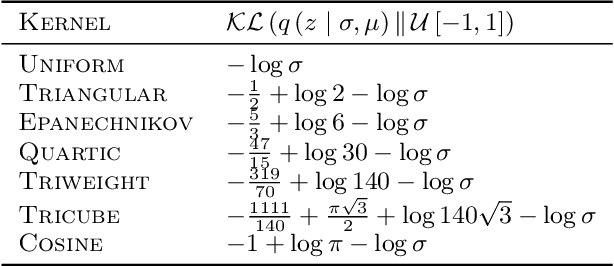
Abstract:Variational autoencoders are prominent generative models for modeling discrete data. However, with flexible decoders, they tend to ignore the latent codes. In this paper, we study a VAE model with a deterministic decoder (DD-VAE) for sequential data that selects the highest-scoring tokens instead of sampling. Deterministic decoding solely relies on latent codes as the only way to produce diverse objects, which improves the structure of the learned manifold. To implement DD-VAE, we propose a new class of bounded support proposal distributions and derive Kullback-Leibler divergence for Gaussian and uniform priors. We also study a continuous relaxation of deterministic decoding objective function and analyze the relation of reconstruction accuracy and relaxation parameters. We demonstrate the performance of DD-VAE on multiple datasets, including molecular generation and optimization problems.
A Prior of a Googol Gaussians: a Tensor Ring Induced Prior for Generative Models
Oct 29, 2019
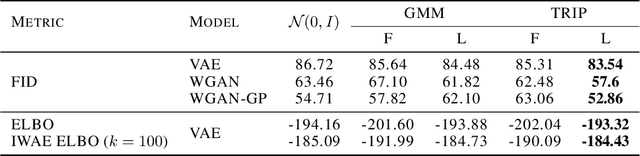


Abstract:Generative models produce realistic objects in many domains, including text, image, video, and audio synthesis. Most popular models---Generative Adversarial Networks (GANs) and Variational Autoencoders (VAEs)---usually employ a standard Gaussian distribution as a prior. Previous works show that the richer family of prior distributions may help to avoid the mode collapse problem in GANs and to improve the evidence lower bound in VAEs. We propose a new family of prior distributions---Tensor Ring Induced Prior (TRIP)---that packs an exponential number of Gaussians into a high-dimensional lattice with a relatively small number of parameters. We show that these priors improve Fr\'echet Inception Distance for GANs and Evidence Lower Bound for VAEs. We also study generative models with TRIP in the conditional generation setup with missing conditions. Altogether, we propose a novel plug-and-play framework for generative models that can be utilized in any GAN and VAE-like architectures.
Molecular Sets (MOSES): A Benchmarking Platform for Molecular Generation Models
Nov 29, 2018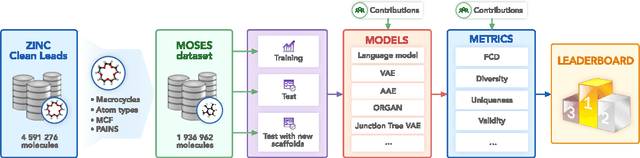
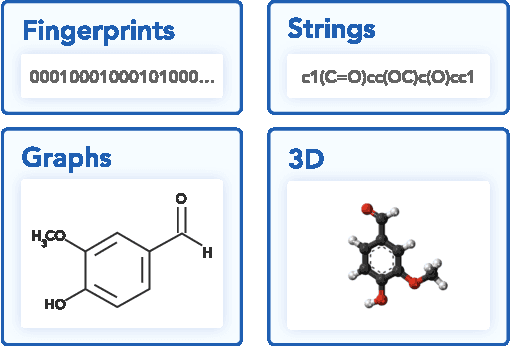

Abstract:Deep generative models such as generative adversarial networks, variational autoencoders, and autoregressive models are rapidly growing in popularity for the discovery of new molecules and materials. In this work, we introduce MOlecular SEtS (MOSES), a benchmarking platform to support research on machine learning for drug discovery. MOSES implements several popular molecular generation models and includes a set of metrics that evaluate the diversity and quality of generated molecules. MOSES is meant to standardize the research on the molecular generation and facilitate the sharing and comparison of new models. Additionally, we provide a large-scale comparison of existing state of the art models and elaborate on current challenges for generative models that might prove fertile ground for new research. Our platform and source code are freely available at https://github.com/molecularsets/
ReSet: Learning Recurrent Dynamic Routing in ResNet-like Neural Networks
Nov 11, 2018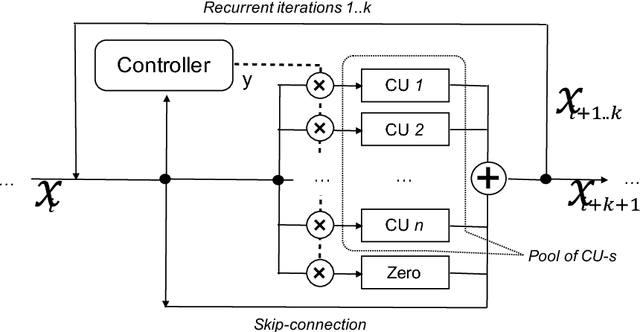
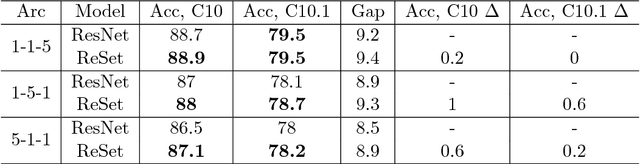
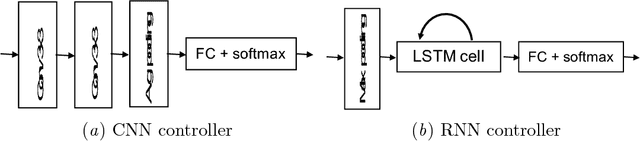
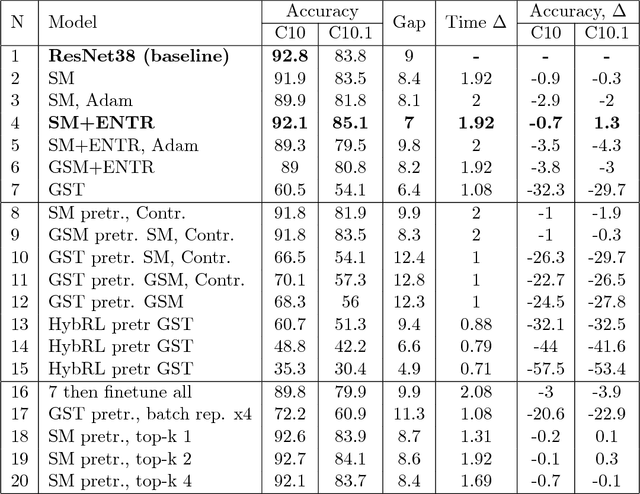
Abstract:Neural Network is a powerful Machine Learning tool that shows outstanding performance in Computer Vision, Natural Language Processing, and Artificial Intelligence. In particular, recently proposed ResNet architecture and its modifications produce state-of-the-art results in image classification problems. ResNet and most of the previously proposed architectures have a fixed structure and apply the same transformation to all input images. In this work, we develop a ResNet-based model that dynamically selects Computational Units (CU) for each input object from a learned set of transformations. Dynamic selection allows the network to learn a sequence of useful transformations and apply only required units to predict the image label. We compare our model to ResNet-38 architecture and achieve better results than the original ResNet on CIFAR-10.1 test set. While examining the produced paths, we discovered that the network learned different routes for images from different classes and similar routes for similar images.
* Published in Proceedings of The 10th Asian Conference on Machine Learning, http://proceedings.mlr.press/v95/kemaev18a.html
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge