Alex Zhavoronkov
When Single Answer Is Not Enough: Rethinking Single-Step Retrosynthesis Benchmarks for LLMs
Feb 03, 2026Abstract:Recent progress has expanded the use of large language models (LLMs) in drug discovery, including synthesis planning. However, objective evaluation of retrosynthesis performance remains limited. Existing benchmarks and metrics typically rely on published synthetic procedures and Top-K accuracy based on single ground-truth, which does not capture the open-ended nature of real-world synthesis planning. We propose a new benchmarking framework for single-step retrosynthesis that evaluates both general-purpose and chemistry-specialized LLMs using ChemCensor, a novel metric for chemical plausibility. By emphasizing plausibility over exact match, this approach better aligns with human synthesis planning practices. We also introduce CREED, a novel dataset comprising millions of ChemCensor-validated reaction records for LLM training, and use it to train a model that improves over the LLM baselines under this benchmark.
BindGPT: A Scalable Framework for 3D Molecular Design via Language Modeling and Reinforcement Learning
Jun 06, 2024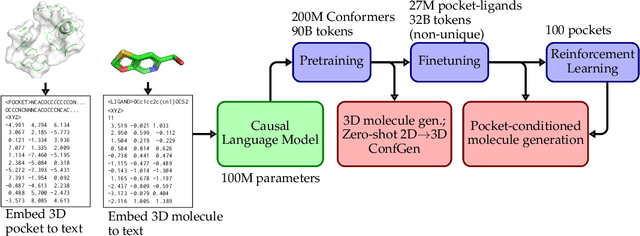
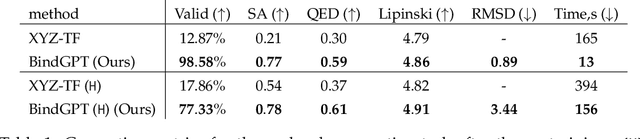
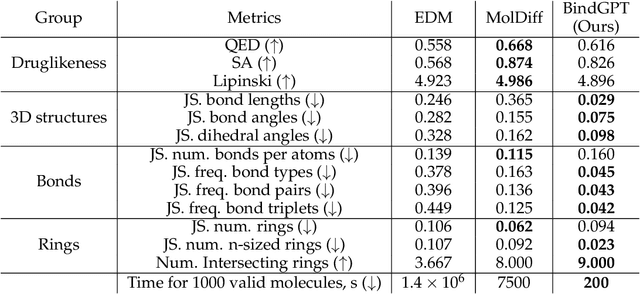
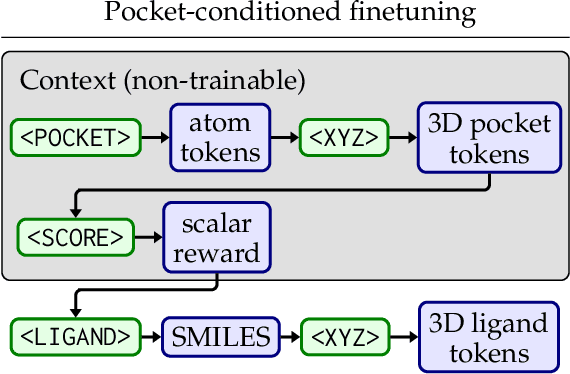
Abstract:Generating novel active molecules for a given protein is an extremely challenging task for generative models that requires an understanding of the complex physical interactions between the molecule and its environment. In this paper, we present a novel generative model, BindGPT which uses a conceptually simple but powerful approach to create 3D molecules within the protein's binding site. Our model produces molecular graphs and conformations jointly, eliminating the need for an extra graph reconstruction step. We pretrain BindGPT on a large-scale dataset and fine-tune it with reinforcement learning using scores from external simulation software. We demonstrate how a single pretrained language model can serve at the same time as a 3D molecular generative model, conformer generator conditioned on the molecular graph, and a pocket-conditioned 3D molecule generator. Notably, the model does not make any representational equivariance assumptions about the domain of generation. We show how such simple conceptual approach combined with pretraining and scaling can perform on par or better than the current best specialized diffusion models, language models, and graph neural networks while being two orders of magnitude cheaper to sample.
Quantum Computing-Enhanced Algorithm Unveils Novel Inhibitors for KRAS
Feb 13, 2024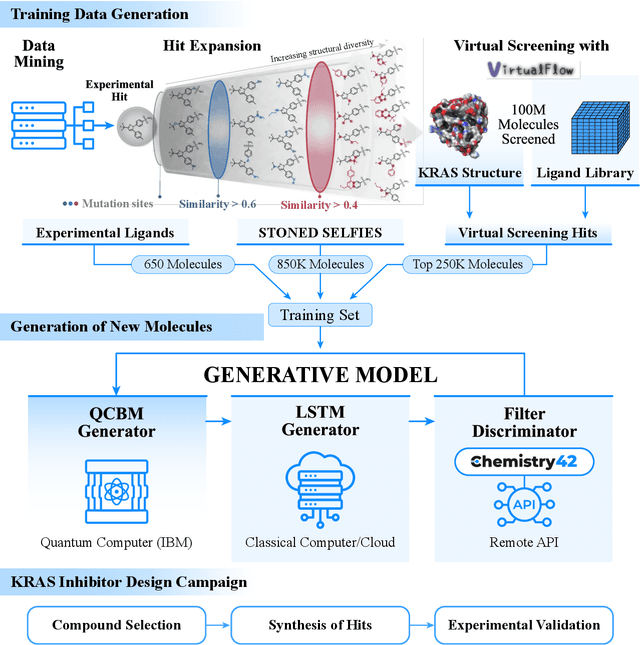
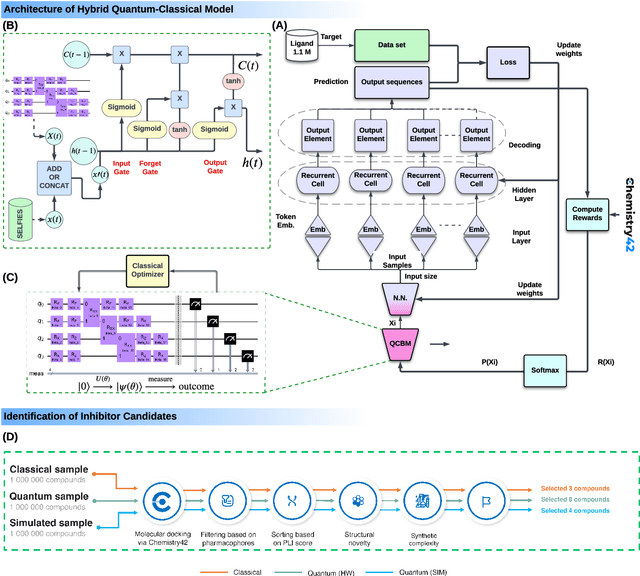
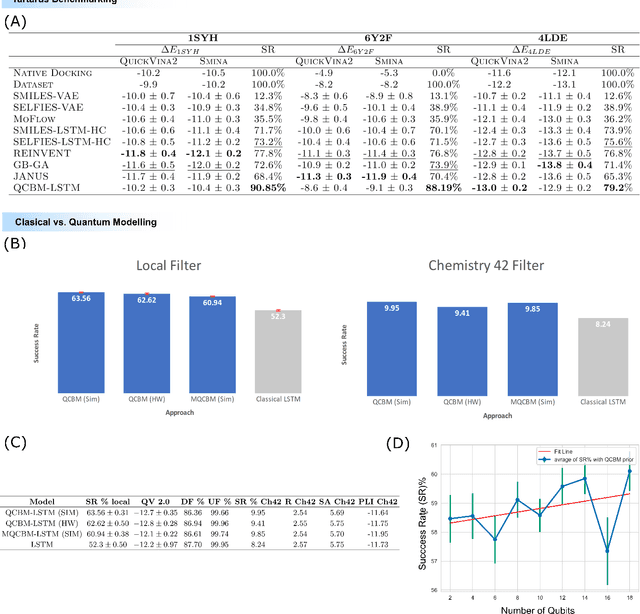
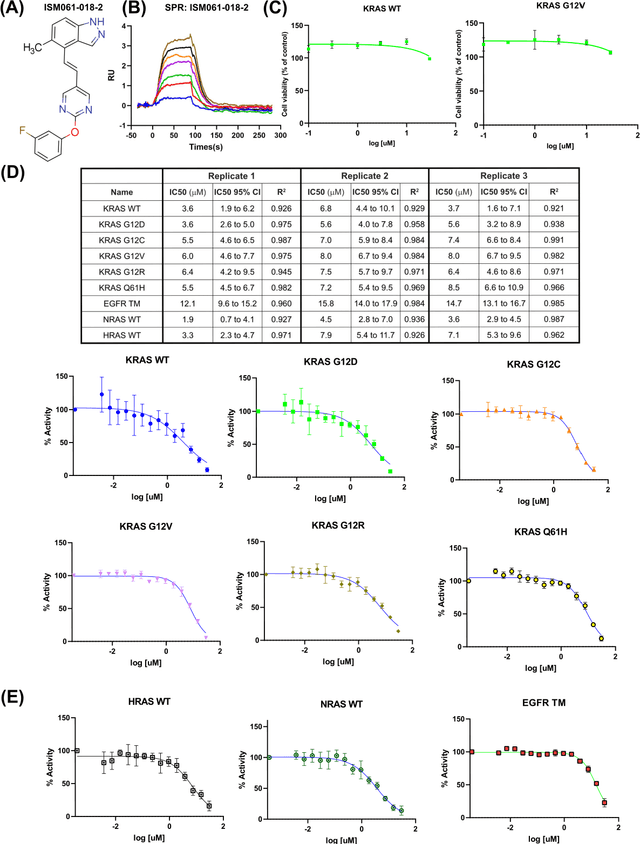
Abstract:The discovery of small molecules with therapeutic potential is a long-standing challenge in chemistry and biology. Researchers have increasingly leveraged novel computational techniques to streamline the drug development process to increase hit rates and reduce the costs associated with bringing a drug to market. To this end, we introduce a quantum-classical generative model that seamlessly integrates the computational power of quantum algorithms trained on a 16-qubit IBM quantum computer with the established reliability of classical methods for designing small molecules. Our hybrid generative model was applied to designing new KRAS inhibitors, a crucial target in cancer therapy. We synthesized 15 promising molecules during our investigation and subjected them to experimental testing to assess their ability to engage with the target. Notably, among these candidates, two molecules, ISM061-018-2 and ISM061-22, each featuring unique scaffolds, stood out by demonstrating effective engagement with KRAS. ISM061-018-2 was identified as a broad-spectrum KRAS inhibitor, exhibiting a binding affinity to KRAS-G12D at $1.4 \mu M$. Concurrently, ISM061-22 exhibited specific mutant selectivity, displaying heightened activity against KRAS G12R and Q61H mutants. To our knowledge, this work shows for the first time the use of a quantum-generative model to yield experimentally confirmed biological hits, showcasing the practical potential of quantum-assisted drug discovery to produce viable therapeutics. Moreover, our findings reveal that the efficacy of distribution learning correlates with the number of qubits utilized, underlining the scalability potential of quantum computing resources. Overall, we anticipate our results to be a stepping stone towards developing more advanced quantum generative models in drug discovery.
nach0: Multimodal Natural and Chemical Languages Foundation Model
Nov 21, 2023



Abstract:Large Language Models (LLMs) have substantially driven scientific progress in various domains, and many papers have demonstrated their ability to tackle complex problems with creative solutions. Our paper introduces a new foundation model, nach0, capable of solving various chemical and biological tasks: biomedical question answering, named entity recognition, molecular generation, molecular synthesis, attributes prediction, and others. nach0 is a multi-domain and multi-task encoder-decoder LLM pre-trained on unlabeled text from scientific literature, patents, and molecule strings to incorporate a range of chemical and linguistic knowledge. We employed instruction tuning, where specific task-related instructions are utilized to fine-tune nach0 for the final set of tasks. To train nach0 effectively, we leverage the NeMo framework, enabling efficient parallel optimization of both base and large model versions. Extensive experiments demonstrate that our model outperforms state-of-the-art baselines on single-domain and cross-domain tasks. Furthermore, it can generate high-quality outputs in molecular and textual formats, showcasing its effectiveness in multi-domain setups.
AlphaFold Accelerates Artificial Intelligence Powered Drug Discovery: Efficient Discovery of a Novel Cyclin-dependent Kinase 20 (CDK20) Small Molecule Inhibitor
Jan 21, 2022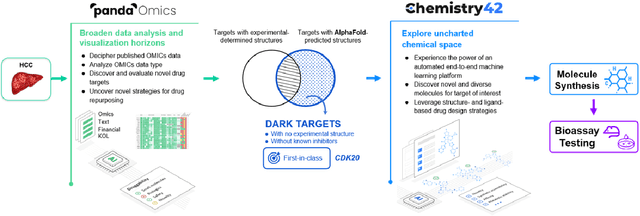
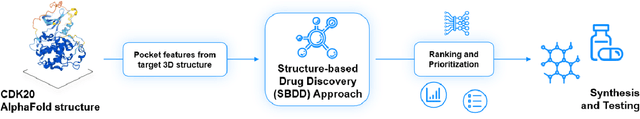
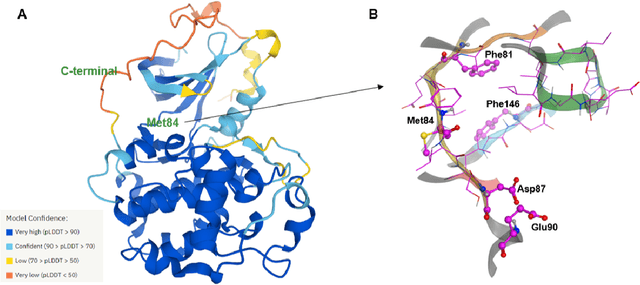
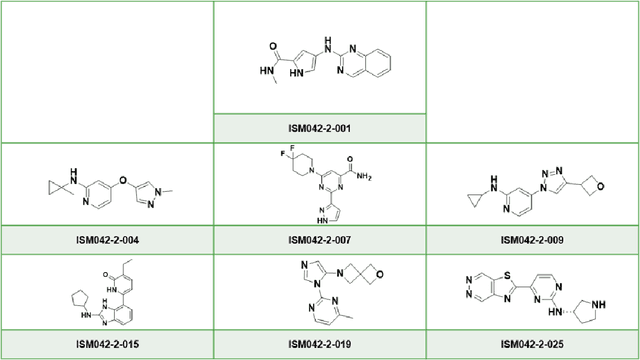
Abstract:The AlphaFold computer program predicted protein structures for the whole human genome, which has been considered as a remarkable breakthrough both in artificial intelligence (AI) application and structural biology. Despite the varying confidence level, these predicted structures still could significantly contribute to the structure-based drug design of novel targets, especially the ones with no or limited structural information. In this work, we successfully applied AlphaFold in our end-to-end AI-powered drug discovery engines constituted of a biocomputational platform PandaOmics and a generative chemistry platform Chemistry42, to identify a first-in-class hit molecule of a novel target without an experimental structure starting from target selection towards hit identification in a cost- and time-efficient manner. PandaOmics provided the targets of interest and Chemistry42 generated the molecules based on the AlphaFold predicted structure, and the selected molecules were synthesized and tested in biological assays. Through this approach, we identified a small molecule hit compound for CDK20 with a Kd value of 8.9 +/- 1.6 uM (n = 4) within 30 days from target selection and after only synthesizing 7 compounds. To the best of our knowledge, this is the first reported small molecule targeting CDK20 and more importantly, this work is the first demonstration of AlphaFold application in the hit identification process in early drug discovery.
Chemistry42: An AI-based platform for de novo molecular design
Jan 22, 2021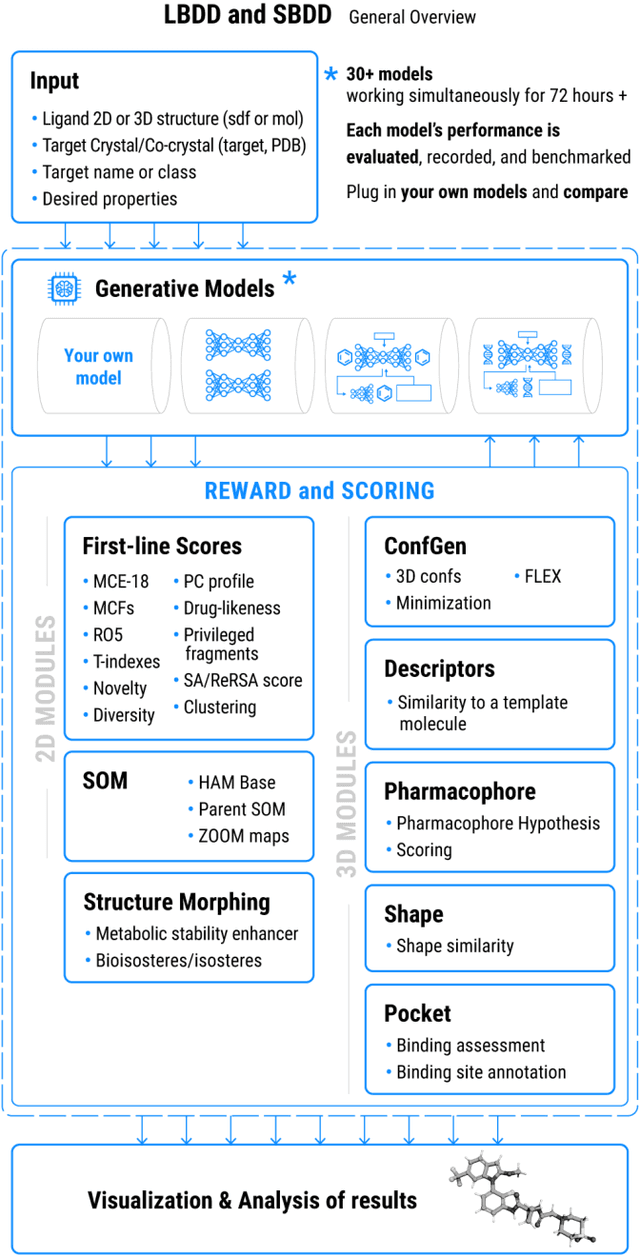
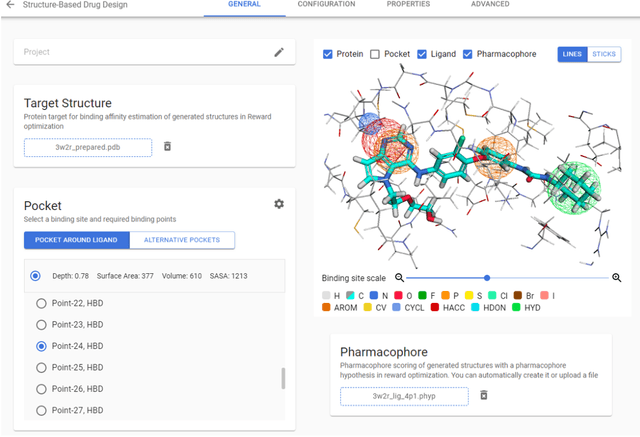
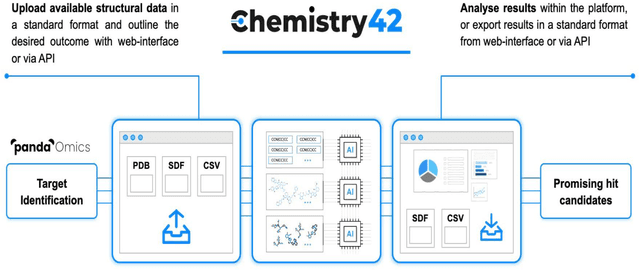
Abstract:Chemistry42 is a software platform for de novo small molecule design that integrates Artificial Intelligence (AI) techniques with computational and medicinal chemistry methods. Chemistry42 is unique in its ability to generate novel molecular structures with predefined properties validated through in vitro and in vivo studies. Chemistry42 is a core component of Insilico Medicine Pharma.ai drug discovery suite that also includes target discovery and multi-omics data analysis (PandaOmics) and clinical trial outcomes predictions (InClinico).
Molecular Sets (MOSES): A Benchmarking Platform for Molecular Generation Models
Nov 29, 2018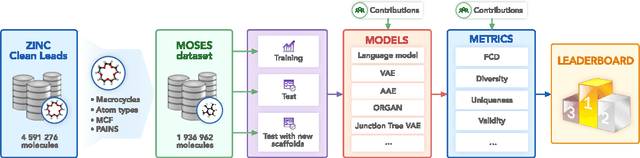

Abstract:Deep generative models such as generative adversarial networks, variational autoencoders, and autoregressive models are rapidly growing in popularity for the discovery of new molecules and materials. In this work, we introduce MOlecular SEtS (MOSES), a benchmarking platform to support research on machine learning for drug discovery. MOSES implements several popular molecular generation models and includes a set of metrics that evaluate the diversity and quality of generated molecules. MOSES is meant to standardize the research on the molecular generation and facilitate the sharing and comparison of new models. Additionally, we provide a large-scale comparison of existing state of the art models and elaborate on current challenges for generative models that might prove fertile ground for new research. Our platform and source code are freely available at https://github.com/molecularsets/
Evaluating race and sex diversity in the world's largest companies using deep neural networks
Jul 09, 2017
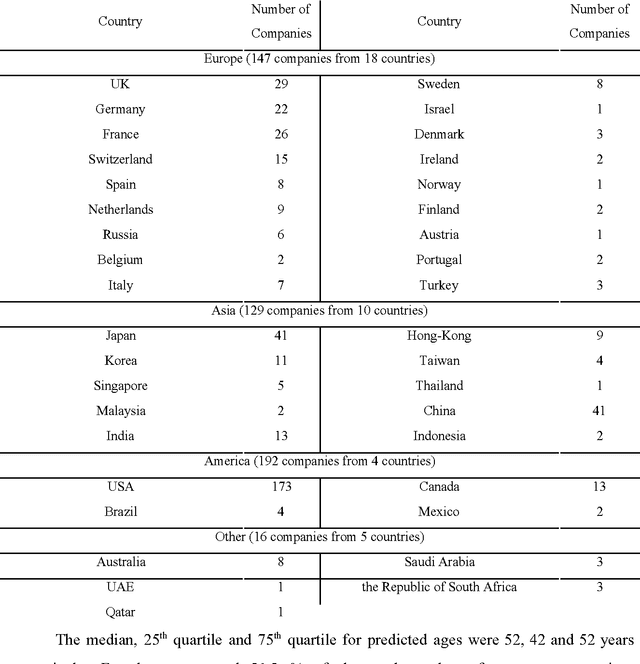

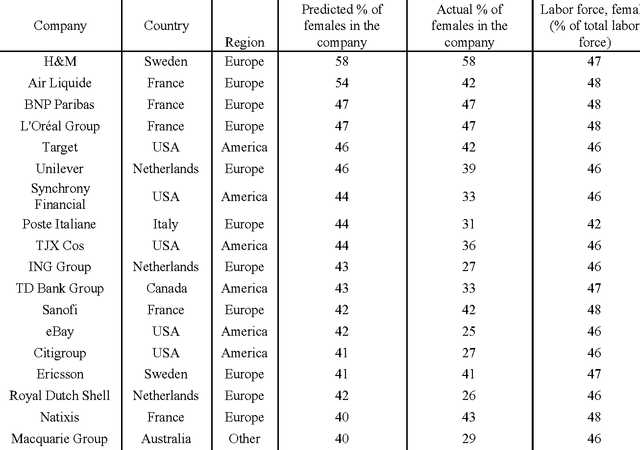
Abstract:Diversity is one of the fundamental properties for the survival of species, populations, and organizations. Recent advances in deep learning allow for the rapid and automatic assessment of organizational diversity and possible discrimination by race, sex, age and other parameters. Automating the process of assessing the organizational diversity using the deep neural networks and eliminating the human factor may provide a set of real-time unbiased reports to all stakeholders. In this pilot study we applied the deep-learned predictors of race and sex to the executive management and board member profiles of the 500 largest companies from the 2016 Forbes Global 2000 list and compared the predicted ratios to the ratios within each company's country of origin and ranked them by the sex-, age- and race- diversity index (DI). While the study has many limitations and no claims are being made concerning the individual companies, it demonstrates a method for the rapid and impartial assessment of organizational diversity using deep neural networks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge