Oktai Tatanov
Automatic Heteronym Resolution Pipeline Using RAD-TTS Aligners
Feb 28, 2023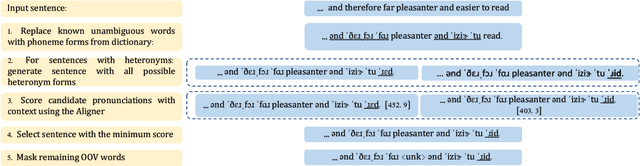



Abstract:Grapheme-to-phoneme (G2P) transduction is part of the standard text-to-speech (TTS) pipeline. However, G2P conversion is difficult for languages that contain heteronyms -- words that have one spelling but can be pronounced in multiple ways. G2P datasets with annotated heteronyms are limited in size and expensive to create, as human labeling remains the primary method for heteronym disambiguation. We propose a RAD-TTS Aligner-based pipeline to automatically disambiguate heteronyms in datasets that contain both audio with text transcripts. The best pronunciation can be chosen by generating all possible candidates for each heteronym and scoring them with an Aligner model. The resulting labels can be used to create training datasets for use in both multi-stage and end-to-end G2P systems.
Mixer-TTS: non-autoregressive, fast and compact text-to-speech model conditioned on language model embeddings
Oct 22, 2021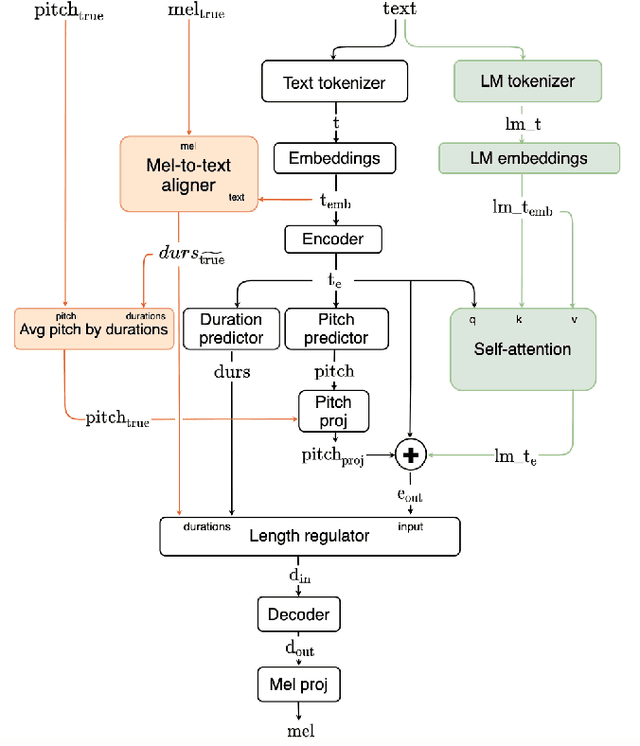


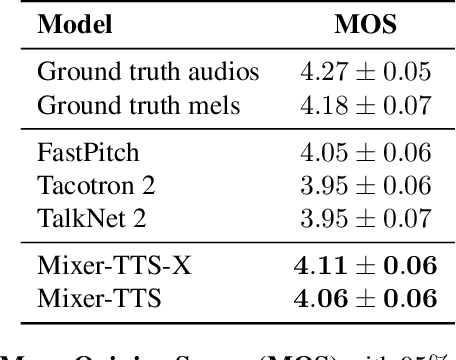
Abstract:This paper describes Mixer-TTS, a non-autoregressive model for mel-spectrogram generation. The model is based on the MLP-Mixer architecture adapted for speech synthesis. The basic Mixer-TTS contains pitch and duration predictors, with the latter being trained with an unsupervised TTS alignment framework. Alongside the basic model, we propose the extended version which additionally uses token embeddings from a pre-trained language model. Basic Mixer-TTS and its extended version achieve a mean opinion score (MOS) of 4.05 and 4.11, respectively, compared to a MOS of 4.27 of original LJSpeech samples. Both versions have a small number of parameters and enable much faster speech synthesis compared to the models with similar quality.
Molecular Sets (MOSES): A Benchmarking Platform for Molecular Generation Models
Nov 29, 2018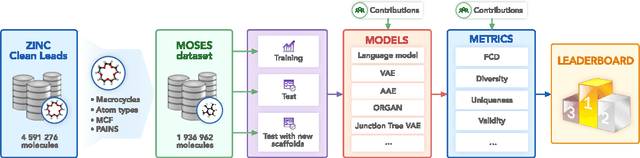
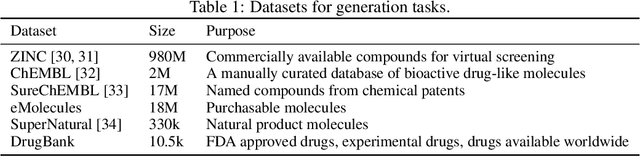
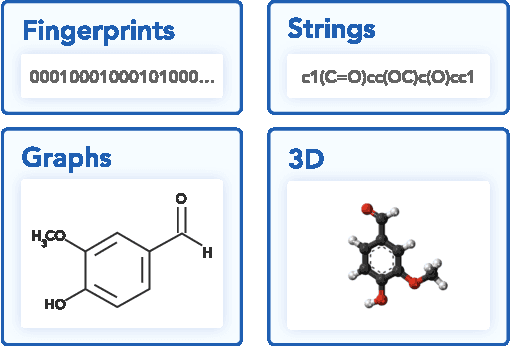

Abstract:Deep generative models such as generative adversarial networks, variational autoencoders, and autoregressive models are rapidly growing in popularity for the discovery of new molecules and materials. In this work, we introduce MOlecular SEtS (MOSES), a benchmarking platform to support research on machine learning for drug discovery. MOSES implements several popular molecular generation models and includes a set of metrics that evaluate the diversity and quality of generated molecules. MOSES is meant to standardize the research on the molecular generation and facilitate the sharing and comparison of new models. Additionally, we provide a large-scale comparison of existing state of the art models and elaborate on current challenges for generative models that might prove fertile ground for new research. Our platform and source code are freely available at https://github.com/molecularsets/
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge